Human CRSP interacts with RNA polymerase II CTD and adopts a specific CTD-bound conformation
- PMID: 12050112
- PMCID: PMC186316
- DOI: 10.1101/gad.987602
Human CRSP interacts with RNA polymerase II CTD and adopts a specific CTD-bound conformation
Abstract
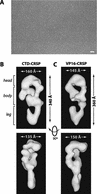
Activation of gene transcription in mammalian cells requires several classes of coactivators that participate in different steps of the activation cascade. Using conventional and affinity chromatography, we have isolated a human coactivator complex that interacts directly with the C-terminal domain (CTD) of RNA polymerase II (Pol II). The CTD-binding complex is structurally and functionally indistinguishable from our previously isolated CRSP coactivator complex. The closely related, but transcriptionally inactive, ARC-L complex failed to interact with the CTD, indicating a significant biochemical difference between CRSP and ARC-L that may, in part, explain their functional divergence. Electron microscopy and three-dimensional single-particle reconstruction reveals a conformation for CTD-CRSP that is structurally distinct from unliganded CRSP or CRSP bound to SREBP-1a, but highly similar to CRSP bound to the VP16 activator. Together, our findings suggest that the human CRSP coactivator functions, at least in part, by mediating activator-dependent recruitment of RNA Pol II via the CTD.
Figures



References
-
- Albright SR, Tjian R. TAFs revisited: More data reveal new twists and confirm old ideas. Gene. 2000;242:1–13. - PubMed
-
- Andel F, Ladurner AG, Inouye C, Tjian R, Nogales E. Three-dimensional structure of the human TFIID-IIA-IIB complex. Science. 1999;286:2153–2156. - PubMed
-
- Boyer TG, Martin MED, Lees E, Ricciardi RP, Berk AJ. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature. 1999;399:276–279. - PubMed
-
- Chiba N, Suldan Z, Freedman LP, Parvin JD. Binding of liganded vitamin D receptor to the vitamin D receptor interacting protein coactivator complex induces interaction with RNA polymerase II holoenzyme. J Biol Chem. 2000;275:10719–10722. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
