Parvovirus initiator protein NS1 and RPA coordinate replication fork progression in a reconstituted DNA replication system
- PMID: 12050365
- PMCID: PMC136255
- DOI: 10.1128/jvi.76.13.6518-6531.2002
Parvovirus initiator protein NS1 and RPA coordinate replication fork progression in a reconstituted DNA replication system
Abstract
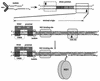
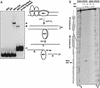
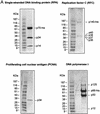
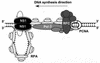
We show here that the DNA helicase activity of the parvoviral initiator protein NS1 is highly directional, binding to the single strand at a recessed 5' end and displacing the other strand while progressing in a 3'-to-5' direction on the bound strand. NS1 and a cellular site-specific DNA binding factor, PIF, also known as glucocorticoid modulating element binding protein, bind to the left-end minimal replication origin of minute virus of mice, forming a ternary complex. In this complex, NS1 is activated to nick one DNA strand, becoming covalently attached to the 5' end of the nick in the process and providing a 3' OH for priming DNA synthesis. In this situation, the helicase activity of NS1 did not displace the nicked strand, but the origin duplex was distorted by the NS1-PIF complex, as assayed by its sensitivity to KMnO(4) oxidation, and a stretch of about 14 nucleotides on both strands of the nicked origin underwent limited unwinding. Addition of Escherichia coli single-stranded DNA binding protein (SSB) did not lead to further unwinding. However, addition of recombinant human single-stranded DNA binding protein (RPA) to the initiation reaction catalyzed extensive unwinding of the nicked origin, suggesting that RPA may be required to form a functional replication fork. Accordingly, the unwinding mediated by NS1 and RPA promoted processive leading-strand synthesis catalyzed by recombinant human DNA polymerase delta, PCNA, and RFC, using the minimal left-end origin cloned in a plasmid as a template. The requirement for RPA, rather than SSB, in the unwinding reaction indicated that specific NS1-RPA protein interactions were formed. NS1 was tested by enzyme-linked immunosorbent assay for binding to two- or three-subunit RPA complexes expressed from recombinant baculoviruses. NS1 efficiently bound each of the baculovirus-expressed complexes, indicating that the small subunit of RPA is not involved in specific NS1 binding. No NS1 interactions were observed with E. coli SSB or other proteins included as controls.
Figures






References
-
- Astell, C. R., Q. Liu, C. E. Harris, J. Brunstein, H. K. Jindal, and P. Tam. 1996. Minute virus of mice cis-acting sequences required for genome replication and the role of the trans-acting viral protein, NS-1. Prog. Nucleic Acid Res. Mol. Biol. 55:245-285. - PubMed
-
- Borowiec, J. A., F. B. Dean, P. A. Bullock, and J. Hurwitz. 1990. Binding and unwinding—how T antigen engages the SV40 origin of DNA replication. Cell J. 60:181-184. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

