P0 of beet Western yellows virus is a suppressor of posttranscriptional gene silencing
- PMID: 12050394
- PMCID: PMC136274
- DOI: 10.1128/jvi.76.13.6815-6824.2002
P0 of beet Western yellows virus is a suppressor of posttranscriptional gene silencing
Retraction in
-
Retraction for Pfeffer et al., P0 of Beet Western Yellows Virus Is a Suppressor of Posttranscriptional Gene Silencing.J Virol. 2017 Mar 15;91(6):e00022-17. doi: 10.1128/JVI.00022-17. J Virol. 2017. PMID: 29611664 Free PMC article. No abstract available.
Abstract
Higher plants employ a homology-dependent RNA-degradation system known as posttranscriptional gene silencing (PTGS) as a defense against virus infection. Several plant viruses are known to encode proteins that can suppress PTGS. Here we show that P0 of beet western yellows virus (BWYV) displays strong silencing suppressor activity in a transient expression assay based upon its ability to inhibit PTGS of green fluorescent protein (GFP) when expressed in agro-infiltrated leaves of Nicotiana benthamiana containing a GFP transgene. PTGS suppressor activity was also observed for the P0s of two other poleroviruses, cucurbit aphid-borne yellows virus and potato leafroll virus. P0 is encoded by the 5'-proximal gene in BWYV RNA but does not accumulate to detectable levels when expressed from the genome-length RNA during infection. The low accumulation of P0 and the resulting low PTGS suppressor activity are in part a consequence of the suboptimal translation initiation context of the P0 start codon in viral RNA, although other factors, probably related to the viral replication process, also play a role. A mutation to optimize the P0 translation initiation efficiency in BWYV RNA was not stable during virus multiplication in planta. Instead, the P0 initiation codon in the progeny was frequently replaced by a less efficient initiation codon such as ACG, GTG, or ATA, indicating that there is selection against overexpression of P0 from the viral genome.
Figures



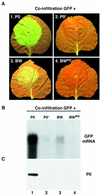
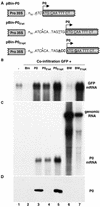

References
-
- Baulcombe, D. C., S. N. Chapman, and S. Santa Cruz. 1995. Jellyfish green fluorescent protein as a reporter for virus infections. Plant J. 7:1045-1053. - PubMed
-
- Béclin, C., R. Berthome, J. C. Palauqui, M. Tepfer, and H. Vaucheret. 1998. Infection of tobacco or Arabidopsis plants by CMV counteracts systemic post-transcriptional silencing of nonviral (trans)genes. Virology 252:313-317. - PubMed
-
- Bosher, J. M., and M. Labouesse. 2000. RNA interference: genetic wand and genetic watchdog. Nat. Cell Biol. 2:E31-E36. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

