Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry
- PMID: 12052880
- PMCID: PMC133885
- DOI: 10.1128/MCB.22.13.4723-4738.2002
Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry
Abstract
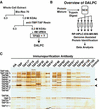
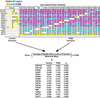
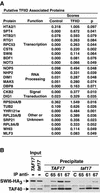
The general transcription factor TFIID is a multisubunit complex of TATA-binding protein (TBP) and 14 distinct TBP-associated factors (TAFs). Although TFIID constituents are required for transcription initiation of most mRNA encoding genes, the mechanism of TFIID action remains unclear. To gain insight into TFIID function, we sought to generate a proteomic catalogue of proteins specifically interacting with TFIID subunits. Toward this end, TFIID was systematically immunopurified by using polyclonal antibodies directed against each subunit, and the constellation of TBP- and TAF-associated proteins was directly identified by coupled multidimensional liquid chromatography and tandem mass spectrometry. A number of novel protein-protein associations were observed, and several were characterized in detail. These interactions include association between TBP and the RSC chromatin remodeling complex, the TAF17p-dependent association of the Swi6p transactivator protein with TFIID, and the identification of three novel subunits of the SAGA acetyltransferase complex, including a putative ubiquitin-specific protease component. Our results provide important new insights into the mechanisms of mRNA gene transcription and demonstrate the feasibility of constructing a complete proteomic interaction map of the eukaryotic transcription apparatus.
Figures




References
-
- Albright, S. R., and R. Tjian. 2000. TAFs revisited: more data reveal new twists and confirm old ideas. Gene 242:1-13. - PubMed
-
- Amerik, A. Y., S. J. Li, and M. Hochstrasser. 2000. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol. Chem. 381:981-992. - PubMed
-
- Angus-Hill, M. L., A. Schlichter, D. Roberts, H. Erdjument-Bromage, P. Tempst, and B. R. Cairns. 2001. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol. Cell 7:741-751. - PubMed
-
- Bell, B., and L. Tora. 1999. Regulation of gene expression by multiple forms of TFIID and other novel TAFII-containing complexes. Exp. Cell Res. 246:11-19. - PubMed
-
- Bell, B., E. Scheer, and L. Tora. 2001. Identification of hTAF(II)80δ links apoptotic signaling pathways to transcription factor TFIID function. Mol. Cell 8:591-600. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
