Nitric oxide-induced homologous recombination in Escherichia coli is promoted by DNA glycosylases
- PMID: 12057944
- PMCID: PMC135131
- DOI: 10.1128/JB.184.13.3501-3507.2002
Nitric oxide-induced homologous recombination in Escherichia coli is promoted by DNA glycosylases
Abstract
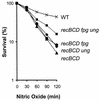
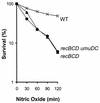
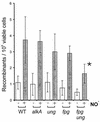
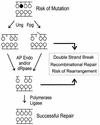
Nitric oxide (NO*) is involved in neurotransmission, inflammation, and many other biological processes. Exposure of cells to NO* leads to DNA damage, including formation of deaminated and oxidized bases. Apurinic/apyrimidinic (AP) endonuclease-deficient cells are sensitive to NO* toxicity, which indicates that base excision repair (BER) intermediates are being generated. Here, we show that AP endonuclease-deficient cells can be protected from NO* toxicity by inactivation of the uracil (Ung) or formamidopyrimidine (Fpg) DNA glycosylases but not by inactivation of a 3-methyladenine (AlkA) DNA glycosylase. These results suggest that Ung and Fpg remove nontoxic NO*-induced base damage to create BER intermediates that are toxic if they are not processed by AP endonucleases. Our next goal was to learn how Ung and Fpg affect susceptibility to homologous recombination. The RecBCD complex is critical for repair of double-strand breaks via homologous recombination. When both Ung and Fpg were inactivated in recBCD cells, survival was significantly enhanced. We infer that both Ung and Fpg create substrates for recombinational repair, which is consistent with the observation that disrupting ung and fpg suppressed NO*-induced recombination. Taken together, a picture emerges in which the action of DNA glycosylases on NO*-induced base damage results in the accumulation of BER intermediates, which in turn can induce homologous recombination. These studies shed light on the underlying mechanism of NO*-induced homologous recombination.
Figures

References
-
- Anderson, D. G., and S. C. Kowalczykowski. 1997. The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a chi-regulated manner. Cell 90:77-86. - PubMed
-
- Burney, S., J. L. Caulfield, J. C. Niles, J. S. Wishnok, and S. R. Tannenbaum. 1999. The chemistry of DNA damage from nitric oxide and peroxynitrite. Mutat. Res. 424:37-49. - PubMed
-
- Burney, S., J. C. Niles, P. C. Dedon, and S. R. Tannenbaum. 1999. DNA damage in deoxynucleosides and oligonucleotides treated with peroxynitrite. Chem. Res. Toxicol. 12:513-520. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

