Differential regulation of twitching motility and elastase production by Vfr in Pseudomonas aeruginosa
- PMID: 12057955
- PMCID: PMC135129
- DOI: 10.1128/JB.184.13.3605-3613.2002
Differential regulation of twitching motility and elastase production by Vfr in Pseudomonas aeruginosa
Abstract
Vfr, a homolog of Escherichia coli cyclic AMP (cAMP) receptor protein, has been shown to regulate quorum sensing, exotoxin A production, and regA transcription in Pseudomonas aeruginosa. We identified a twitching motility-defective mutant that carries a transposon insertion in vfr and confirmed that vfr is required for twitching motility by construction of an independent allelic deletion-replacement mutant of vfr that exhibited the same phenotype, as well as by the restoration of normal twitching motility by complementation of these mutants with wild-type vfr. Vfr-null mutants exhibited severely reduced twitching motility with barely detectable levels of type IV pili, as well as loss of elastase production and altered pyocyanin production. We also identified reduced-twitching variants of quorum-sensing mutants (PAK lasI::Tc) with a spontaneous deletion in vfr (S. A. Beatson, C. B. Whitchurch, A. B. T. Semmler, and J. S. Mattick, J. Bacteriol., 184:3598-3604, 2002), the net result of which was the loss of five residues (EQERS) from the putative cAMP-binding pocket of Vfr. This allele (VfrDeltaEQERS) was capable of restoring elastase and pyocyanin production to wild-type levels in vfr-null mutants but not their defects in twitching motility. Furthermore, structural analysis of Vfr and VfrDeltaEQERS in relation to E. coli CRP suggests that Vfr is capable of binding both cAMP and cyclic GMP whereas VfrDeltaEQERS is only capable of responding to cAMP. We suggest that Vfr controls twitching motility and quorum sensing via independent pathways in response to these different signals, bound by the same cyclic nucleotide monophosphate-binding pocket.
Figures


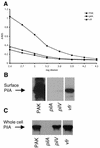

References
-
- Alm, R. A., and J. S. Mattick. 1997. Genes involved in the biogenesis and function of type-4 fimbriae in Pseudomonas aeruginosa. Gene 192:89-98. - PubMed
-
- Alm, R. A., and J. S. Mattick. 1995. Identification of a gene, pilV, required for type 4 fimbrial biogenesis in Pseudomonas aeruginosa whose product possesses a prepilin-like leader sequence. Mol. Microbiol. 16:485-496. - PubMed
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

