Presence of large deletions in kindreds with autism
- PMID: 12058345
- PMCID: PMC384967
- DOI: 10.1086/341291
Presence of large deletions in kindreds with autism
Abstract
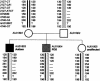
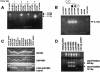
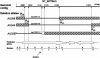
Autism is caused, in part, by inheritance of multiple interacting susceptibility alleles. To identify these inherited factors, linkage analysis of multiplex families is being performed on a sample of 105 families with two or more affected sibs. Segregation patterns of short tandem repeat polymorphic markers from four chromosomes revealed null alleles at four marker sites in 12 families that were the result of deletions ranging in size from 5 to >260 kb. In one family, a deletion at marker D7S630 was complex, with two segments deleted (37 kb and 18 kb) and two retained (2,836 bp and 38 bp). Three families had deletions at D7S517, with each family having a different deletion (96 kb, 183 kb, and >69 kb). Another three families had deletions at D8S264, again with each family having a different deletion, ranging in size from <5.9 kb to >260 kb. At a fourth marker, D8S272, a 192-kb deletion was found in five families. Unrelated subjects and additional families without autism were screened for deletions at these four sites. Families screened included 40 families from Centre d'Etude du Polymorphisme Humaine and 28 families affected with learning disabilities. Unrelated samples were 299 elderly control subjects, 121 younger control subjects, and 248 subjects with Alzheimer disease. The deletion allele at D8S272 was found in all populations screened. For the other three sites, no additional deletions were identified in any of the groups without autism. Thus, these deletions appear to be specific to autism kindreds and are potential autism-susceptibility alleles. An alternative hypothesis is that autism-susceptibility alleles elsewhere cause the deletions detected here, possibly by inducing errors during meiosis.
Figures

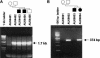



References
Electronic-Database Information
-
- dbSNP Database, http://www.ncbi.nlm.nih.gov/SNP/ (for data on individual SNPs)
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for sequence and SNP information for genomic contigs NT_017168.5 [accession number GI:15298273], NT_007784.5 [accession number GI:15299808], NT_008104.5 [accession number GI:15300207], and NT_008087.5 [accession number GI:15300158])
-
- Genetic Epidemiology of Complex Traits, http://www.stat.washington.edu/thompson/Genepi/ (for the computer program Loki)
-
- Seqhelp, http://biocommons.bcc.washington.edu/services/psoftware.seqhelp/seqhelpi... (for identification of repeat sequences using RepeatMaster, ProcessRepeats, and Repbase)
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for autism [MIM 209850], FMR1 [MIM 309550], TS [MIM 191100], PWS [MIM 176270], AS [MIM 105830], HOXA1 [MIM 142955], HOXB1 [MIM 142968], EN-2 [MIM 131310], SLC6A4 [MIM 182138], RELN [MIM 600514], WNT2 [MIM 147870], VIPR2 [MIM 601970], RAY1 [MIM 600833], GABRB3 [MIM 137192], VCFS [MIM 192430], MYOM2 [MIM 603509], LOAD [MIM 104300], HSD [MIM 235730], SCZD [MIM 181500], XPC [MIM 278720], and PML [MIM 102578])
References
-
- American Psychiatric Association (1994) Diagnostic and statistical manual of mental disorders (4th ed). American Psychiatric Association, Washington, DC, pp 65–78
-
- Asano E, Kuivaniemi H, Huq M, Tromp G, Behen M, Rothermel R, Herron J, Chugani DC (2001) A study of novel polymorphisms in the upstream region of vasoactive intestinal peptide receptor type 2 gene in autism. J Child Neurol 16:357–363 - PubMed
-
- Ashley-Koch A, Wolpert CM, Menold MM, Zaeem L, Basu S, Donnelly SL, Ravan SA, Powell CM, Qumsiyeh MB, Aylsworth AS, Vance JM, Gilbert JR, Wright HH, Abramson RK, Delong GR, Cuccaro ML, Pericak-Vance MAJ (1999) Genetic studies of autistic disorder and chromosome 7. Genomics 61:227–236 - PubMed
-
- Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M (1995) Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med 25:63–77 - PubMed
-
- Baker P, Piven J, Schwartz S, Patil S (1994) Brief report: duplication of chromosome 15q11-13 in two individuals with autistic disorder. J Autism Dev Disord 24:529–535 - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

