Heterozygous submicroscopic inversions involving olfactory receptor-gene clusters mediate the recurrent t(4;8)(p16;p23) translocation
- PMID: 12058347
- PMCID: PMC379160
- DOI: 10.1086/341610
Heterozygous submicroscopic inversions involving olfactory receptor-gene clusters mediate the recurrent t(4;8)(p16;p23) translocation
Abstract
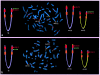
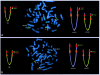
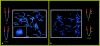
The t(4;8)(p16;p23) translocation, in either the balanced form or the unbalanced form, has been reported several times. Taking into consideration the fact that this translocation may be undetected in routine cytogenetics, we find that it may be the most frequent translocation after t(11q;22q), which is the most common reciprocal translocation in humans. Case subjects with der(4) have the Wolf-Hirschhorn syndrome, whereas case subjects with der(8) show a milder spectrum of dysmorphic features. Two pairs of the many olfactory receptor (OR)-gene clusters are located close to each other, on both 4p16 and 8p23. Previously, we demonstrated that an inversion polymorphism of the OR region at 8p23 plays a crucial role in the generation of chromosomal imbalances through unusual meiotic exchanges. These findings prompted us to investigate whether OR-related inversion polymorphisms at 4p16 and 8p23 might also be involved in the origin of the t(4;8)(p16;p23) translocation. In seven case subjects (five of whom both represented de novo cases and were of maternal origin), including individuals with unbalanced and balanced translocations, we demonstrated that the breakpoints fell within the 4p and 8p OR-gene clusters. FISH experiments with appropriate bacterial-artificial-chromosome probes detected heterozygous submicroscopic inversions of both 4p and 8p regions in all the five mothers of the de novo case subjects. Heterozygous inversions on 4p16 and 8p23 were detected in 12.5% and 26% of control subjects, respectively, whereas 2.5% of them were scored as doubly heterozygous. These novel data emphasize the importance of segmental duplications and large-scale genomic polymorphisms in the evolution and pathology of the human genome.
Figures

References
Electronic-Database Information
-
- BACPAC Resources, http://www.chori.org/bacpac/
-
- Celera Publication Site, http://publication.celera.com/cds/login.cfm
-
- Database of Human Olfactory Receptor Genes, A, http://bioinformatics.weizmann.ac.il/HORDE/humanGenes/
-
- Entrez Genome View, http://www.ncbi.nlm.nih.gov/cgi-bin/Entrez/hum_srch?chr=hum_chr.inf&query (for Homo sapiens genome view, build 29)
References
-
- Bondeson ML, Dahl N, Malmgren H, Kleijer WJ, Tonnesen T, Carlberg BM, Pettersson U (1995) Inversion of the IDS gene resulting from recombination with IDS-related sequences is a common cause of the Hunter syndrome. Hum Mol Genet 4:615–621 - PubMed
-
- Eichler EE (2001) Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends Genet 17:661–669 - PubMed
-
- Emanuel BS, Shaikh TH (2001) Segmental duplications: an “expanding” role in genomic instability and disease. Nat Rev Genet 2:791–800 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

