Regulated HIV-2 RNA dimerization by means of alternative RNA conformations
- PMID: 12060681
- PMCID: PMC117293
- DOI: 10.1093/nar/gkf381
Regulated HIV-2 RNA dimerization by means of alternative RNA conformations
Abstract
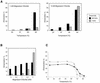
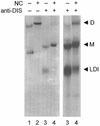
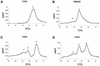
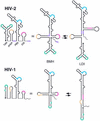
The dimer initiation site (DIS) hairpin of the HIV-2 untranslated leader RNA mediates in vitro dimerization through 'loop-loop kissing' of a loop-exposed palindrome sequence. Premature RNA dimerization must be prevented during the retroviral life cycle. A regulatory mechanism has been proposed for the HIV-1 leader RNA that can adopt an alternative conformation in which the DIS motif is effectively masked by long-distance base pairing with upstream leader sequences. We now report that HIV-2 RNA dimerization is also regulated. Sequestering of the DIS motif by base pairing interactions with downstream leader sequences mediates a switch to a dimerization-impaired conformation. The existence of two alternative conformations of the HIV-2 leader RNA is supported by UV melting experiments. Furthermore, the equilibrium between the two conformations can be shifted by annealing of antisense oligonucleotides or by deletion of certain leader regions. These measures have a profound impact on the dimerization properties of the transcript, demonstrating a mutual exclusivity between the alternative conformation and dimerization, similar to what has been described for the HIV-1 leader. The overall resemblance in regulation of HIV-1 and HIV-2 RNA dimerization suggests that a similar mechanism may be operating in other lentiviruses and perhaps all retroviridae.
Figures
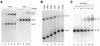
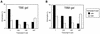

References
-
- Coffin J.M. (1990) Retroviridae and their replication. In Fields,B.N. and Knipe,D.M. (eds), Virology. Raven Press Ltd, NY.
-
- Greatorex J. and Lever,A. (1998) Retroviral RNA dimer linkage. J. Gen. Virol., 79, 2877–2882. - PubMed
-
- Darlix J.L., Gabus,C., Nugeyre,M.T., Clavel,F. and Barre-Sinoussi,F. (1990) Cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J. Mol. Biol., 216, 689–699. - PubMed
-
- Gotte M., Li,X. and Wainberg,M.A. (1999) HIV-1 reverse transcription: a brief overview focused on structure-function relationships among molecules involved in initiation of the reaction. Arch. Biochem. Biophys., 365, 199–210. - PubMed

