High-throughput genotyping of single nucleotide polymorphisms using new biplex invader technology
- PMID: 12060691
- PMCID: PMC117295
- DOI: 10.1093/nar/gnf052
High-throughput genotyping of single nucleotide polymorphisms using new biplex invader technology
Abstract
The feasibility of large-scale genome-wide association studies of complex human disorders depends on the availability of accurate and efficient genotyping methods for single nucleotide polymorphisms (SNPs). We describe a new platform of the invader assay, a biplex assay, where both alleles are interrogated in a single reaction tube. The assay was evaluated on over 50 different SNPs, with over 20 SNPs genotyped in study cohorts of over 1500 individuals. We assessed the usefulness of the new platform in high-throughput genotyping and compared its accuracy to genotyping results obtained by the traditional monoplex invader assay, TaqMan genotyping and sequencing data. We present representative data for two SNPs in different genes (CD36 and protein tyrosine phosphatase 1beta) from a study cohort comprising over 1500 individuals with high or low-normal blood pressure. In this high-throughput application, the biplex invader assay is very accurate, with an error rate of <0.3% and a failure rate of 1.64%. The set-up of the assay is highly automated, facilitating the processing of large numbers of samples simultaneously. We present new analysis tools for the assignment of genotypes that further improve genotyping success. The biplex invader assay with its automated set-up and analysis offers a new efficient high-throughput genotyping platform that is suitable for association studies in large study cohorts.
Figures
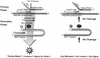


References
-
- Sachidanandam R., Weissman,D., Schmidt,S.C., Kakol,J.M., Stein,L.D., Marth,G., Sherry,S., Mullikin,J.C., Mortimore,B.J., Willey,D.L. et al. (2001) A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature, 409, 928–933. - PubMed
-
- Risch N. and Merikangas,K. (1996) The future of genetic studies of complex human diseases. Science, 273, 1516–1517. - PubMed
-
- Kwok P.-Y. (2001) Genetic association by whole-genome analysis? Science, 294, 1669–1670. - PubMed
-
- Kwok P.-Y. (2000) High-throughput genotyping assay approaches. Pharmacogenomics, 1, 95–100. - PubMed
-
- Livak K.J., Marmaro,J. and Todd,J.A. (1995) Towards fully automated genome-wide polymorphism screening. Nature Genet., 9, 341–342. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

