A universal method to produce in vitro transcripts with homogeneous 3' ends
- PMID: 12060694
- PMCID: PMC117298
- DOI: 10.1093/nar/gnf055
A universal method to produce in vitro transcripts with homogeneous 3' ends
Abstract
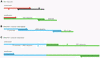
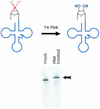
A method is described that allows a general drawback of in vitro transcription assays to be overcome: RNA polymerases tend to add extra nucleotides to the RNA 3' end that are not encoded in the linearized DNA template. Furthermore, these polymerases show a considerable rate of premature termination close to the RNA's 3' end. These features lead to a decreased yield of full-length transcripts and often make it difficult to determine and isolate the correctly transcribed full-length RNA. The hammerhead ribozyme is frequently used in cis to cleave off these extra nucleotides. However, the upstream sequence requirements of this ribozyme restrict its general usability. In contrast, the hepatitis delta virus ribozyme has no such requirements and can therefore be applied to any RNA sequence in cis. Due to the catalytic activity of the ribozyme, the desired transcript is released as an RNA molecule with a homogeneous 3' end. The resulting 2',3'-cyclo-phosphate group of the released RNA can be easily and efficiently removed by T4 polynucleotide kinase treatment. The presented method can be applied for virtually any sequence to be transcribed and is therefore superior to other ribozyme strategies, suggesting possible applications in every field where transcripts with homogeneous 3' ends are required.
Figures


References
-
- Chamberlin M.J. and Ryan,T. (1982) Bacteriophage DNA-dependent RNA polymerases. In Boyer,P.D. (ed.), The Enzymes. Academic Press, New York, Vol. 15B, pp. 87–109.
-
- Krieg P.A. and Melton,D.A. (1987) In vitro RNA synthesis with SP6 RNA polymerase. Methods Enzymol., 155, 397–415. - PubMed
-
- Milligan J.F. and Uhlenbeck,O.C. (1989) Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol., 180, 51–62. - PubMed
-
- Draper D.E., White,S.A. and Kean,J.M. (1988) Preparation of specific ribosomal RNA fragments. Methods Enzymol., 164, 221–237. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

