Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification
- PMID: 12060695
- PMCID: PMC117299
- DOI: 10.1093/nar/gnf056
Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification
Abstract
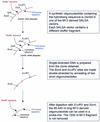
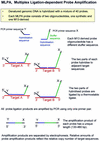
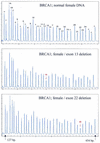
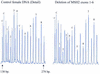
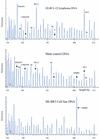
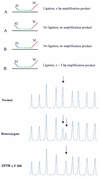
We describe a new method for relative quantification of 40 different DNA sequences in an easy to perform reaction requiring only 20 ng of human DNA. Applications shown of this multiplex ligation-dependent probe amplification (MLPA) technique include the detection of exon deletions and duplications in the human BRCA1, MSH2 and MLH1 genes, detection of trisomies such as Down's syndrome, characterisation of chromosomal aberrations in cell lines and tumour samples and SNP/mutation detection. Relative quantification of mRNAs by MLPA will be described elsewhere. In MLPA, not sample nucleic acids but probes added to the samples are amplified and quantified. Amplification of probes by PCR depends on the presence of probe target sequences in the sample. Each probe consists of two oligonucleotides, one synthetic and one M13 derived, that hybridise to adjacent sites of the target sequence. Such hybridised probe oligonucleotides are ligated, permitting subsequent amplification. All ligated probes have identical end sequences, permitting simultaneous PCR amplification using only one primer pair. Each probe gives rise to an amplification product of unique size between 130 and 480 bp. Probe target sequences are small (50-70 nt). The prerequisite of a ligation reaction provides the opportunity to discriminate single nucleotide differences.
Figures


References
-
- Petrij-Bosch A., Peelen,T., van Vliet,M., van Eijk,R., Olmer,R., Drusedau,M., Hogervorst,F.B., Hageman,S., Arts,P.J., Ligtenberg,M.J. et al. (1997) BRCAI genomic deletions are major founder mutations in Dutch breast cancer patients. Nature Genet., 17, 341–345. - PubMed
-
- Wijnen J., van der Klift,H., Vasen,H., Khan,P.M., Menko,F., Tops,C., Meijers Heijboer,H., Lindhout,D., Moller,P. and Fodde,R. (1998) MSH2 genomic deletions are a frequent cause of HNPCC. Nature Genet., 20, 326–328. - PubMed
-
- Kauraniemi P., Barlund,M., Monni,O. and Kallioniemi,A. (2001) New amplified and highly expressed genes discovered in the ERBB2 amplicon in breast cancer by cDNA microarrays. Cancer Res., 61, 8235–8240. - PubMed
-
- Leyland-Jones B. and Smith,I. (2001) Role of herceptin in primary breast cancer. Oncology, 61 (Suppl. 2), 83–91. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

