Methylation of histone H3 Lys 4 in coding regions of active genes
- PMID: 12060701
- PMCID: PMC124361
- DOI: 10.1073/pnas.082249499
Methylation of histone H3 Lys 4 in coding regions of active genes
Abstract
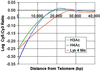
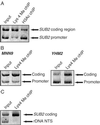
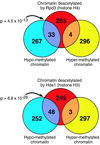
Posttranslational modifications of histone tails regulate chromatin structure and transcription. Here we present global analyses of histone acetylation and histone H3 Lys 4 methylation patterns in yeast. We observe a significant correlation between acetylation of histones H3 and H4 in promoter regions and transcriptional activity. In contrast, we find that dimethylation of histone H3 Lys 4 in coding regions correlates with transcriptional activity. The histone methyltransferase Set1 is required to maintain expression of these active, promoter-acetylated, and coding region-methylated genes. Global comparisons reveal that genomic regions deacetylated by the yeast enzymes Rpd3 and Hda1 overlap extensively with Lys 4 hypo- but not hypermethylated regions. In the context of recent studies showing that Lys 4 methylation precludes histone deacetylase recruitment, we conclude that Set1 facilitates transcription, in part, by protecting active coding regions from deacetylation.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

