Integration of Runx and Smad regulatory signals at transcriptionally active subnuclear sites
- PMID: 12060751
- PMCID: PMC123018
- DOI: 10.1073/pnas.112664499
Integration of Runx and Smad regulatory signals at transcriptionally active subnuclear sites
Abstract
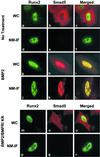
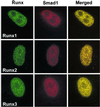
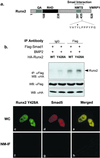
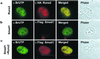
Runx factors control lineage commitment and are transcriptional effectors of Smad signaling. Genetic defects in these pathways interfere with normal development. The in situ localization of Runx and Smad proteins must impact the mechanisms by which these proteins function together in gene regulation. We show that the integration of Runx and Smad signals is mediated by in situ interactions at specific foci within the nucleus. Activated Smads are directed to these subnuclear foci only in the presence of Runx proteins. Smad-Runx complexes are associated in situ with the nuclear matrix, and this association requires the intranuclear targeting signal of Runx factors. The convergence of Smad and Runx proteins at these sites supports transcription as reflected by BrUTP labeling and functional cooperativity between the proteins. Thus, Runx-mediated intranuclear targeting of Smads is critical for the integration of two distinct pathways essential for fetal development.
Figures

References
-
- Massague J. Annu Rev Biochem. 1998;67:753–791. - PubMed
-
- Finidori J. Vitam Horm. 2000;59:71–97. - PubMed
-
- Ihle J N. Curr Opin Cell Biol. 2001;13:211–217. - PubMed
-
- Akiyama T. Cytokine Growth Factor Rev. 2000;11:273–282. - PubMed
-
- Merriman H L, van Wijnen A J, Hiebert S, Bidwell J P, Fey E, Lian J, Stein J, Stein G S. Biochemistry. 1995;34:13125–13132. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

