Modulation of c-myc, max, and mad gene expression during neural differentiation of embryonic stem cells by all-trans-retinoic acid
- PMID: 12064575
- PMCID: PMC5977512
Modulation of c-myc, max, and mad gene expression during neural differentiation of embryonic stem cells by all-trans-retinoic acid
Abstract
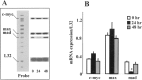
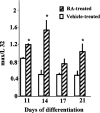
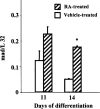
c-Myc regulates cellular proliferation, differentiation, and apoptosis. Temporal expression of c-Myc during all-trans-retinoic acid (RA)-mediated neural differentiation in murine embryonic stem cell (ES) was investigated. Correlation to the modulation of dimerizing partners Max and Mad that may influence c-Myc signaling and transcription regulation was elucidated for the first time in these cells. In RA-treated cells, increase in c-myc mRNA was detected by reverse transcriptase polymerase chain reaction on days 11 and 14 of differentiation compared with the vehicle-treated controls. The results were further corroborated by ribonuclease protection assay (RPA). Western blots revealed an increase in c-Myc protein only on day 14 of differentiation in RA-treated cells. Increases in max and mad gene transcription detected by RPA at times of elevated c-Myc in RA-treated ES cells suggest that a transient increase in c-Myc protein expression may influence differential dimerization of Myc partners needed for signaling in the neural differentiation of ES cells.
Figures




Similar articles
-
Mad3 and Mad4: novel Max-interacting transcriptional repressors that suppress c-myc dependent transformation and are expressed during neural and epidermal differentiation.EMBO J. 1995 Nov 15;14(22):5646-59. doi: 10.1002/j.1460-2075.1995.tb00252.x. EMBO J. 1995. PMID: 8521822 Free PMC article.
-
Retinoic acid-induced growth arrest and differentiation of neuroblastoma cells are counteracted by N-myc and enhanced by max overexpressions.Oncogene. 1996 Jan 18;12(2):457-62. Oncogene. 1996. PMID: 8570225
-
A switch from Myc:Max to Mad:Max heterocomplexes accompanies monocyte/macrophage differentiation.Genes Dev. 1993 Nov;7(11):2110-9. doi: 10.1101/gad.7.11.2110. Genes Dev. 1993. PMID: 8224841
-
The Max transcription factor network: involvement of Mad in differentiation and an approach to identification of target genes.Cold Spring Harb Symp Quant Biol. 1994;59:109-16. doi: 10.1101/sqb.1994.059.01.014. Cold Spring Harb Symp Quant Biol. 1994. PMID: 7587059 Review.
-
Myc-Max-Mad: a transcription factor network controlling cell cycle progression, differentiation and death.Curr Opin Genet Dev. 1994 Feb;4(1):102-8. doi: 10.1016/0959-437x(94)90098-1. Curr Opin Genet Dev. 1994. PMID: 8193530 Review.
Cited by
-
Analysis of the Polymorphisms and Expression Levels of the BCL2, BAX and c-MYC Genes in Patients with Ovarian Cancer.Int J Mol Sci. 2023 Nov 14;24(22):16309. doi: 10.3390/ijms242216309. Int J Mol Sci. 2023. PMID: 38003498 Free PMC article.
-
The RNA-Binding Protein HuD Regulates Alternative Splicing and Alternative Polyadenylation in the Mouse Neocortex.Molecules. 2021 May 11;26(10):2836. doi: 10.3390/molecules26102836. Molecules. 2021. PMID: 34064652 Free PMC article.
-
Genomic structure, promoter activity, and developmental expression of the mouse homologue of the Machado-Joseph disease (MJD) gene.Genomics. 2004 Aug;84(2):361-73. doi: 10.1016/j.ygeno.2004.02.012. Genomics. 2004. PMID: 15233999 Free PMC article.
-
Expression profile of differentiating serotonin neurons derived from rhesus embryonic stem cells and comparison to adult serotonin neurons.Gene Expr Patterns. 2009 Feb;9(2):94-108. doi: 10.1016/j.gep.2008.10.002. Epub 2008 Nov 1. Gene Expr Patterns. 2009. PMID: 18996226 Free PMC article.
-
Human iPSC-derived neural stem cells displaying radial glia signature exhibit long-term safety in mice.Nat Commun. 2024 Nov 1;15(1):9433. doi: 10.1038/s41467-024-53613-7. Nat Commun. 2024. PMID: 39487141 Free PMC article.
References
-
- Amati B.; Alevizopoulos K.; Vlach J. Myc and the cell cycle. Front. Biosci. 3:D250–D268; 1998. - PubMed
-
- Amati B.; Land H. Myc-Max-Mad: A transcription factor network controlling cell cycle progression, differentiation and death. Curr. Opin. Genet. Dev. 4:102–108; 1994. - PubMed
-
- Amati B.; Dalton S.; Brooks M. W.; Littlewood T. D.; Evan G. I.; Land H. Transcriptional activation by the human c-Myc oncoprotein in yeast requires interaction with Max. Nature 359:423–426; 1992. - PubMed
-
- Ayer D. E.; Eisenman R. N. A switch from Myc:Max to Mad:Max heterocomplexes accompanies monocyte/macrophage differentiation. Genes Dev. 7:2110–2119; 1993. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
