Crystal structure of vesicular stomatitis virus matrix protein
- PMID: 12065402
- PMCID: PMC126044
- DOI: 10.1093/emboj/cdf284
Crystal structure of vesicular stomatitis virus matrix protein
Abstract
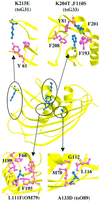
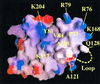
The vesicular stomatitis virus (VSV) matrix protein (M) interacts with cellular membranes, self-associates and plays a major role in virus assembly and budding. We present the crystallographic structure, determined at 1.96 A resolution, of a soluble thermolysin resistant core of VSV M. The fold is a new fold shared by the other vesiculovirus matrix proteins. The structure accounts for the loss of stability of M temperature-sensitive mutants deficient in budding, and reveals a flexible loop protruding from the globular core that is important for self-assembly. Membrane floatation shows that, together with the M lysine-rich N-terminal peptide, a second domain of the protein is involved in membrane binding. Indeed, the structure reveals a hydrophobic surface located close to the hydrophobic loop and surrounded by conserved basic residues that may constitute this domain. Lastly, comparison of the negative-stranded virus matrix proteins with retrovirus Gag proteins suggests that the flexible link between their major membrane binding domain and the rest of the structure is a common feature shared by these proteins involved in budding and virus assembly.
Figures



Similar articles
-
Assembly functions of vesicular stomatitis virus matrix protein are not disrupted by mutations at major sites of phosphorylation.Virology. 1995 Feb 1;206(2):894-903. doi: 10.1006/viro.1995.1012. Virology. 1995. PMID: 7856102
-
Tracking the Fate of Genetically Distinct Vesicular Stomatitis Virus Matrix Proteins Highlights the Role for Late Domains in Assembly.J Virol. 2015 Dec;89(23):11750-60. doi: 10.1128/JVI.01371-15. Epub 2015 Sep 2. J Virol. 2015. PMID: 26339059 Free PMC article.
-
Cleavage of vesicular stomatitis virus matrix protein prevents self-association and leads to crystallization.Virology. 2001 Sep 30;288(2):308-14. doi: 10.1006/viro.2001.1062. Virology. 2001. PMID: 11601902
-
The role of vesicular stomatitis virus matrix protein in inhibition of host-directed gene expression is genetically separable from its function in virus assembly.J Virol. 1993 Aug;67(8):4814-21. doi: 10.1128/JVI.67.8.4814-4821.1993. J Virol. 1993. PMID: 8392615 Free PMC article.
-
A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding.J Virol. 1999 Apr;73(4):2921-9. doi: 10.1128/JVI.73.4.2921-2929.1999. J Virol. 1999. PMID: 10074141 Free PMC article.
Cited by
-
The Functional Oligomeric State of Tegument Protein GP41 Is Essential for Baculovirus Budded Virion and Occlusion-Derived Virion Assembly.J Virol. 2018 May 29;92(12):e02083-17. doi: 10.1128/JVI.02083-17. Print 2018 Jun 15. J Virol. 2018. PMID: 29643237 Free PMC article.
-
Characterization of the Interaction between the Matrix Protein of Vesicular Stomatitis Virus and the Immunoproteasome Subunit LMP2.J Virol. 2015 Nov;89(21):11019-29. doi: 10.1128/JVI.01753-15. Epub 2015 Aug 26. J Virol. 2015. PMID: 26311888 Free PMC article.
-
IFN-beta-induced alteration of VSV protein phosphorylation in neuronal cells.Viral Immunol. 2009 Dec;22(6):353-69. doi: 10.1089/vim.2009.0057. Viral Immunol. 2009. PMID: 19951173 Free PMC article.
-
Atomic model of vesicular stomatitis virus and mechanism of assembly.Nat Commun. 2022 Oct 10;13(1):5980. doi: 10.1038/s41467-022-33664-4. Nat Commun. 2022. PMID: 36216930 Free PMC article.
-
Vesiculoviral matrix (M) protein occupies nucleic acid binding site at nucleoporin pair (Rae1 • Nup98).Proc Natl Acad Sci U S A. 2014 Jun 24;111(25):9127-32. doi: 10.1073/pnas.1409076111. Epub 2014 Jun 9. Proc Natl Acad Sci U S A. 2014. PMID: 24927547 Free PMC article.
References
-
- Abrahams J.P. and Leslie,A.G.W. (1996) Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D, 52, 30–42. - PubMed
-
- Arzt S., Baudin,F., Barge,A., Timmins,P., Burmeister,W.P. and Ruigrok,R.W. (2001) Combined results from solution studies on intact influenza virus M1 protein and from a new crystal form of its N-terminal domain show that M1 is an elongated monomer. Virology, 279, 439–446. - PubMed
-
- Barge A., Gagnon,J., Chaffotte,A., Timmins,P., Langowsky,J., Ruigrok,R. and Gaudin,Y. (1996) Rod-like shape of vesicular stomatitis virus matrix protein. Virology, 219, 465–470. - PubMed
-
- Baudin F., Petit,I., Weissenhorn,W. and Ruigrok,R.W. (2001) In vitro dissection of the membrane and RNP binding activities of influenza virus M1 protein. Virology, 281, 102–108. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

