Homolog interaction during meiotic prophase I in Arabidopsis requires the SOLO DANCERS gene encoding a novel cyclin-like protein
- PMID: 12065421
- PMCID: PMC126045
- DOI: 10.1093/emboj/cdf285
Homolog interaction during meiotic prophase I in Arabidopsis requires the SOLO DANCERS gene encoding a novel cyclin-like protein
Abstract
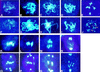
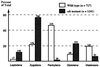
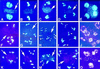
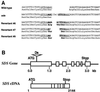
Interactions between homologs in meiotic prophase I, such as recombination and synapsis, are critical for proper homolog segregation and involve the coordination of several parallel events. However, few regulatory genes have been identified; in particular, it is not clear what roles the proteins similar to the mitotic cell cycle regulators might play during meiotic prophase I. We describe here the isolation and characterization of a new Arabidopsis mutant called solo dancers that exhibits a severe defect in homolog synapsis, recombination and bivalent formation in meiotic prophase I, subsequently resulting in seemingly random chromosome distribution and formation of abnormal meiotic products. We further demonstrate that the mutation affects a meiosis-specific gene encoding a novel protein of 578 amino acid residues with up to 31% amino acid sequence identity to known cyclins in the C-terminal portion. These results argue strongly that homolog interactions during meiotic prophase I require a novel meiosis-specific cyclin in Arabidopsis.
Figures





References
-
- Alexander M.P. (1969) Differential staining of aborted and nonaborted pollen. Stain Technol., 44, 117–122. - PubMed
-
- Andrews B. and Measday,V. (1998) The cyclin family of budding yeast: abundant use of a good idea. Trends Genet., 14, 66–72. - PubMed
-
- Armstrong S.J. and Jones,G.H. (2001) Female meiosis in wild-type Arabidopsis thialiana and two meiotic mutants. Sex. Plant Reprod., 13, 177–183.
-
- Ashley T. and Plug,A. (1998) Caught in the act: deducing meiotic function from protein immunolocalization. In Handel,M.A. (ed.), Meiosis and Gametogenesis. Academic Press, San Diego, Vol. 37, pp. 201–239. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

