Candida albicans hyphal formation and the expression of the Efg1-regulated proteinases Sap4 to Sap6 are required for the invasion of parenchymal organs
- PMID: 12065511
- PMCID: PMC128044
- DOI: 10.1128/IAI.70.7.3689-3700.2002
Candida albicans hyphal formation and the expression of the Efg1-regulated proteinases Sap4 to Sap6 are required for the invasion of parenchymal organs
Abstract
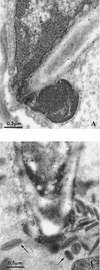
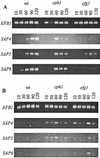
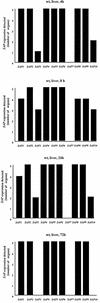
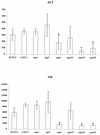
The ability to change between yeast and hyphal cells (dimorphism) is known to be a virulence property of the human pathogen Candida albicans. The pathogenesis of disseminated candidosis involves adhesion and penetration of hyphal cells from a colonized mucosal site to internal organs. Parenchymal organs, such as the liver and pancreas, are invaded by C. albicans wild-type hyphal cells between 4 and 24 h after intraperitoneal (i.p.) infection of mice. In contrast, a hypha-deficient mutant lacking the transcription factor Efg1 was not able to invade or damage these organs. To investigate whether this was due to the inability to undergo the dimorphic transition or due to the lack of hypha-associated factors, we investigated the role of secreted aspartic proteinases during tissue invasion and their association with the different morphologies of C. albicans. Wild-type cells expressed a distinct pattern of SAP genes during i.p. infections. Within the first 72 h after infection, SAP1, SAP2, SAP4, SAP5, SAP6, and SAP9 were the most commonly expressed proteinase genes. Sap1 to Sap3 antigens were found on yeast and hyphal cells, while Sap4 to Sap6 antigens were predominantly found on hyphal cells in close contact with host cells, in particular, eosinophilic leukocytes. Mutants lacking EFG1 had either noticeably reduced or higher expressed levels of SAP4 to SAP6 transcripts in vitro depending on the culture conditions. During infection, efg1 mutants had a strongly reduced ability to produce hyphae, which was associated with reduced levels of SAP4 to SAP6 transcripts. Mutants lacking SAP1 to SAP3 had invasive properties indistinguishable from those of wild-type cells. In contrast, a triple mutant lacking SAP4 to SAP6 showed strongly reduced invasiveness but still produced hyphal cells. When the tissue damage of liver and pancreas caused by single sap4, sap5, and sap6 and double sap4 and -6, sap5 and -6, and sap4 and -5 double mutants was compared to the damage caused by wild-type cells, all mutants which lacked functional SAP6 showed significantly reduced tissue damage. These data demonstrate that strains which produce hyphal cells but lack hypha-associated proteinases, particularly that encoded by SAP6, are less invasive. In addition, it can be concluded that the reduced virulence of hypha-deficient mutants is not only due to the inability to form hyphae but also due to modified expression of the SAP genes normally associated with the hyphal morphology.
Figures



Similar articles
-
Reduced expression of the hyphal-independent Candida albicans proteinase genes SAP1 and SAP3 in the efg1 mutant is associated with attenuated virulence during infection of oral epithelium.J Med Microbiol. 2003 Aug;52(Pt 8):623-632. doi: 10.1099/jmm.0.05125-0. J Med Microbiol. 2003. PMID: 12867554
-
The role of secreted aspartyl proteinases in Candida albicans keratitis.Invest Ophthalmol Vis Sci. 2007 Aug;48(8):3559-65. doi: 10.1167/iovs.07-0114. Invest Ophthalmol Vis Sci. 2007. PMID: 17652724
-
The secreted aspartyl proteinases Sap1 and Sap2 cause tissue damage in an in vitro model of vaginal candidiasis based on reconstituted human vaginal epithelium.Infect Immun. 2003 Jun;71(6):3227-34. doi: 10.1128/IAI.71.6.3227-3234.2003. Infect Immun. 2003. PMID: 12761103 Free PMC article.
-
[What functions do six different genes for secretory proteinases have in Candida albicans?].Mycoses. 1998;41 Suppl 1:47-50. doi: 10.1111/j.1439-0507.1998.tb00583.x. Mycoses. 1998. PMID: 9717386 Review. German.
-
Candida albicans secreted aspartyl proteinases.Curr Top Med Mycol. 1996 Dec;7(1):55-69. Curr Top Med Mycol. 1996. PMID: 9504059 Review.
Cited by
-
Structural Insights into the Interactions of Candidal Enolase with Human Vitronectin, Fibronectin and Plasminogen.Int J Mol Sci. 2020 Oct 22;21(21):7843. doi: 10.3390/ijms21217843. Int J Mol Sci. 2020. PMID: 33105833 Free PMC article.
-
The glycosylphosphatidylinositol-anchored protease Sap9 modulates the interaction of Candida albicans with human neutrophils.Infect Immun. 2009 Dec;77(12):5216-24. doi: 10.1128/IAI.00723-09. Epub 2009 Oct 5. Infect Immun. 2009. PMID: 19805528 Free PMC article.
-
ERG3 and ERG11 genes are critical for the pathogenesis of Candida albicans during the oral mucosal infection.Int J Oral Sci. 2018 Mar 16;10(2):9. doi: 10.1038/s41368-018-0013-2. Int J Oral Sci. 2018. PMID: 29555898 Free PMC article.
-
Effects of surface reaction-type pre-reacted glass ionomer on oral biofilm formation of Streptococcus gordonii.Odontology. 2016 Sep;104(3):310-7. doi: 10.1007/s10266-015-0217-2. Epub 2015 Aug 30. Odontology. 2016. PMID: 26319990
-
Evolutionary Selection on Barrier Activity: Bar1 Is an Aspartyl Protease with Novel Substrate Specificity.mBio. 2015 Nov 24;6(6):e01604-15. doi: 10.1128/mBio.01604-15. mBio. 2015. PMID: 26604258 Free PMC article.
References
-
- Borg-von Zepelin, M., S. Beggah, K. Boggian, D. Sanglard, and M. Monod. 1998. The expression of the secreted aspartic proteinases Sap4 to Sap6 from Candida albicans in murine macrophages. Mol. Microbiol. 28:543-554. - PubMed
-
- Brown, A. J., and N. A. Gow. 1999. Regulatory networks controlling Candida albicans morphogenesis. Trends Microbiol. 7:333-338. - PubMed
-
- Buffo, J., M. A. Herman, and D. R. Soll. 1984. A characterization of pH-regulated dimorphism in Candida albicans. Mycopathologia 85:21-30. - PubMed
-
- Cutler, J. E. 1991. Putative virulence factors of Candida albicans. Annu. Rev. Microbiol. 45:187-218. - PubMed
-
- De Bernardis, F., S. Arancia, L. Morelli, B. Hube, D. Sanglard, W. Schäfer, and A. Cassone. 1999. Evidence that members of the secretory aspartic proteinase gene family, in particular SAP2, are virulence factors for Candida vaginitis. J. Infect. Dis. 179:201-208. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

