NPSN11 is a cell plate-associated SNARE protein that interacts with the syntaxin KNOLLE
- PMID: 12068098
- PMCID: PMC161670
- DOI: 10.1104/pp.003970
NPSN11 is a cell plate-associated SNARE protein that interacts with the syntaxin KNOLLE
Abstract
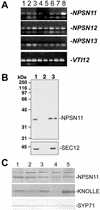
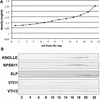
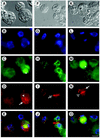
SNAREs are important components of the vesicle trafficking machinery in eukaryotic cells. In plants, SNAREs have been found to play a variety of roles in the development and physiology of the whole organism. Here, we describe the identification and characterization of a novel plant-specific SNARE, NPSN11, a member of a closely related small gene family in Arabidopsis. NSPN11 is highly expressed in actively dividing cells. In a subcellular fractionation experiment, NSPN11 cofractionates with the cytokinesis-specific syntaxin, KNOLLE, which is required for the formation of the cell plate. By immunofluorescence microscopy, NSPN11 was localized to the cell plate in dividing cells. Consistent with the localization studies, NSPN11 was found to interact with KNOLLE. Our results suggest that NPSN11 is another component of the membrane trafficking and fusion machinery involved in cell plate formation.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

