The branched-chain amino acid transaminase gene family in Arabidopsis encodes plastid and mitochondrial proteins
- PMID: 12068099
- PMCID: PMC161671
- DOI: 10.1104/pp.001602
The branched-chain amino acid transaminase gene family in Arabidopsis encodes plastid and mitochondrial proteins
Abstract
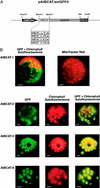
Branched-chain amino acid transaminases (BCATs) play a crucial role in the metabolism of leucine, isoleucine, and valine. They catalyze the last step of the synthesis and/or the initial step of the degradation of this class of amino acids. In Arabidopsis, seven putative BCAT genes are identified by their similarity to their counterparts from other organisms. We have now cloned the respective cDNA sequences of six of these genes. The deduced amino acid sequences show between 47.5% and 84.1% identity to each other and about 30% to the homologous enzymes from yeast (Saccharomyces cerevisiae) and mammals. In addition, many amino acids in crucial positions as determined by crystallographic analyses of BCATs from Escherichia coli and human (Homo sapiens) are conserved in the AtBCATs. Complementation of a yeast Deltabat1/Deltabat2 double knockout strain revealed that five AtBCATs can function as BCATs in vivo. Transient expression of BCAT:green fluorescent protein fusion proteins in tobacco (Nicotiana tabacum) protoplasts shows that three isoenzymes are imported into chloroplasts (AtBCAT-2, -3, and -5), whereas a single enzyme is directed into mitochondria (AtBCAT-1).
Figures


References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Aubert S, Alban C, Bligny R, Douce R. Induction of beta-methylcrotonyl-coenzyme A carboxylase in higher plant cells during carbohydrate starvation: evidence for a role of MCCase in leucine catabolism. FEBS Lett. 1996;383:175–180. - PubMed
-
- Bledsoe RK, Dawson PA, Hutson SM. Cloning of the rat and human mitochondrial branched chain aminotransferases (BCATm) Biochim Biophys Acta. 1997;1339:9–13. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

