RASPBERRY3 gene encodes a novel protein important for embryo development
- PMID: 12068112
- PMCID: PMC161694
- DOI: 10.1104/pp.004010
RASPBERRY3 gene encodes a novel protein important for embryo development
Erratum in
- Plant Physiol 2002 Oct;130(2):1073. Harada John H [corrected to Harada John J]
Abstract
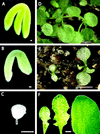
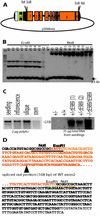
We identified a new gene that is interrupted by T-DNA in an Arabidopsis embryo mutant called raspberry3. raspberry3 has "raspberry-like" cellular protuberances with an enlarged suspensor characteristic of other raspberry embryo mutants, and is arrested morphologically at the globular stage of embryo development. The predicted RASPBERRY3 protein has domains found in proteins present in prokaryotes and algae chloroplasts. Computer prediction analysis suggests that the RASPBERRY3protein may be localized in the chloroplast. Complementation analysis supports the possibility that the RASPBERRY3 protein may be involved in chloroplast development. Our experiments demonstrate the important role of the chloroplast, directly or indirectly, in embryo morphogenesis and development.
Figures




References
-
- Albert S, Despres B, Guilleminot J, Bechtold N, Pelletier G, Delseny M, Devic M. The EMB 506 gene encodes a novel ankyrin repeat containing protein that is essential for the normal development of Arabidopsis embryos. Plant J. 1999;17:169–179. - PubMed
-
- Altmann T, Felix G, Jessop A, Kauschmann A, Uwer U, Peña-Cortés H, Willmitzer L. Ac/Ds transposon mutagenesis in Arabidopsis thaliana: mutant spectrum and frequency of Ds insertion mutants. Mol Gen Genet. 1995;247:646–652. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

