Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor
- PMID: 12068113
- PMCID: PMC161695
- DOI: 10.1104/pp.001057
Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor
Abstract
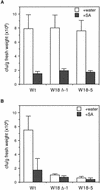
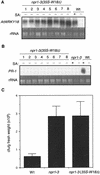
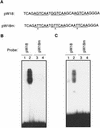
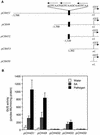
AtWRKY18 is a pathogen- and salicylic acid-induced Arabidopsis transcription factor containing the plant-specific WRKY zinc finger DNA-binding motif. In the present study, we have transformed Arabidopsis plants with AtWRKY18 under control of the cauliflower mosaic virus 35S promoter. Surprisingly, transgenic plants expressing high levels of AtWRKY18 were stunted in growth. When expressed at moderate levels, AtWRKY18 potentiated developmentally regulated defense responses in transgenic plants without causing substantial negative effects on plant growth. As they grew from seedling to mature stages, transgenic AtWRKY18 plant showed marked increase in the expression of pathogenesis-related genes and resistance to the bacterial pathogen Pseudomonas syringae, whereas wild-type plants exhibited little enhancement in these defense responses. Potentiation of developmentally regulated defense responses by AtWRKY18 was not associated with enhanced biosynthesis of salicylic acid but required the disease resistance regulatory protein NPR1/NIM1. Thus, AtWRKY18 can positively modulate defense-related gene expression and disease resistance. To study the regulated expression of AtWRKY18, we have identified a cluster of WRKY binding sites in the promoter of the gene and demonstrated that they acted as negative regulatory elements for the inducible expression of AtWRKY18. These negative cis-acting elements may prevent overexpression of AtWRKY18 during the activation of plant defense responses that could be detrimental to plant growth as inferred from the transgenic plants ectopically expressing the transgene.
Figures




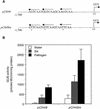
References
-
- Bechtold N, Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol. 1998;82:259–266. - PubMed
-
- Cameron RK, Dixon RA, Lamb CJ. Biologically induced systemic acquired resistance in Arabidopsis thaliana. Plant J. 1994;5:715–725.
-
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell. 1997;88:57–63. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

