Evidence for a Ustilago maydis steroid 5alpha-reductase by functional expression in Arabidopsis det2-1 mutants
- PMID: 12068114
- PMCID: PMC161696
- DOI: 10.1104/pp.001016
Evidence for a Ustilago maydis steroid 5alpha-reductase by functional expression in Arabidopsis det2-1 mutants
Abstract
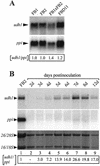
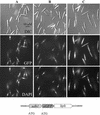
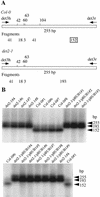
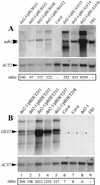
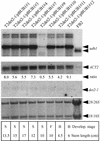
We have identified a gene (udh1) in the basidiomycete Ustilago maydis that is induced during the parasitic interaction with its host plant maize (Zea mays). udh1 encodes a protein with high similarity to mammalian and plant 5alpha-steroid reductases. Udh1 differs from those of known 5alpha-steroid reductases by six additional domains, partially predicted to be membrane-spanning. A fusion protein of Udh1 and the green fluorescent protein provided evidence for endoplasmic reticulum localization in U. maydis. The function of the Udh1 protein was demonstrated by complementing Arabidopsis det2-1 mutants, which display a dwarf phenotype due to a mutation in the 5alpha-steroid reductase encoding DET2 gene. det2-1 mutant plants expressing either the udh1 or the DET2 gene controlled by the cauliflower mosaic virus 35S promoter differed from wild-type Columbia plants by accelerated stem growth, flower and seed development and a reduction in size and number of rosette leaves. The accelerated growth phenotype of udh1 transgenic plants was stably inherited and was favored under reduced light conditions. Truncation of the N-terminal 70 amino acids of the Udh1 protein abolished the ability to restore growth in det2-1 plants. Our results demonstrate the existence of a 5alpha-steroid reductase encoding gene in fungi and suggest a common ancestor between fungal, plant, and mammalian proteins.
Figures





References
-
- Aichinger C. Identifizierung pflanzenabhängig-regulierter Gene in Ustilago maydis. PhD thesis. Munich, Germany: LMU München; 2000.
-
- Altmann T. A tale of dwarfs and drugs: brassinosteroids to the rescue. Trends Genet. 1998;14:490–495. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

