Gene expression profile of HIV-1 Tat expressing cells: a close interplay between proliferative and differentiation signals
- PMID: 12069692
- PMCID: PMC116586
- DOI: 10.1186/1471-2091-3-14
Gene expression profile of HIV-1 Tat expressing cells: a close interplay between proliferative and differentiation signals
Abstract
Background: Expression profiling holds great promise for rapid host genome functional analysis. It is plausible that host expression profiling in an infection could serve as a universal phenotype in virally infected cells. Here, we describe the effect of one of the most critical viral activators, Tat, in HIV-1 infected and Tat expressing cells. We utilized microarray analysis from uninfected, latently HIV-1 infected cells, as well as cells that express Tat, to decipher some of the cellular changes associated with this viral activator.
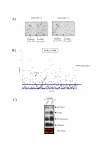
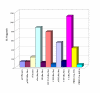
Results: Utilizing uninfected, HIV-1 latently infected cells, and Tat expressing cells, we observed that most of the cellular host genes in Tat expressing cells were down-regulated. The down-regulation in Tat expressing cells is most apparent on cellular receptors that have intrinsic receptor tyrosine kinase (RTK) activity and signal transduction members that mediate RTK function, including Ras-Raf-MEK pathway. Co-activators of transcription, such as p300/CBP and SRC-1, which mediate gene expression related to hormone receptor genes, were also found to be down-regulated. Down-regulation of receptors may allow latent HIV-1 infected cells to either hide from the immune system or avoid extracellular differentiation signals. Some of the genes that were up-regulated included co-receptors for HIV-1 entry, translation machinery, and cell cycle regulatory proteins.
Conclusions: We have demonstrated, through a microarray approach, that HIV-1 Tat is able to regulate many cellular genes that are involved in cell signaling, translation and ultimately control the host proliferative and differentiation signals.
Figures


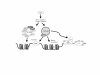

Similar articles
-
Effect of SWI/SNF chromatin remodeling complex on HIV-1 Tat activated transcription.Retrovirology. 2006 Aug 7;3:48. doi: 10.1186/1742-4690-3-48. Retrovirology. 2006. PMID: 16893449 Free PMC article.
-
Human immunodeficiency virus type 1 Tat increases the expression of cleavage and polyadenylation specificity factor 73-kilodalton subunit modulating cellular and viral expression.J Virol. 2004 Jul;78(13):6846-54. doi: 10.1128/JVI.78.13.6846-6854.2004. J Virol. 2004. PMID: 15194760 Free PMC article.
-
Down-modulation of TCR/CD3 surface complexes after HIV-1 infection is associated with differential expression of the viral regulatory genes.Eur J Immunol. 2001 Apr;31(4):969-79. doi: 10.1002/1521-4141(200104)31:4<969::aid-immu969>3.0.co;2-2. Eur J Immunol. 2001. PMID: 11298321
-
Multiple effects of HIV-1 trans-activator protein on the pathogenesis of HIV-1 infection.Eur J Clin Invest. 2004 Jan;34(1):57-66. doi: 10.1111/j.1365-2362.2004.01282.x. Eur J Clin Invest. 2004. PMID: 14984439 Review.
-
Multiple modes of transcriptional regulation by the HIV-1 Tat transactivator.IUBMB Life. 2001 Mar;51(3):175-81. doi: 10.1080/152165401753544241. IUBMB Life. 2001. PMID: 11547919 Review.
Cited by
-
Host cell gene expression during human immunodeficiency virus type 1 latency and reactivation and effects of targeting genes that are differentially expressed in viral latency.J Virol. 2004 Sep;78(17):9458-73. doi: 10.1128/JVI.78.17.9458-9473.2004. J Virol. 2004. PMID: 15308739 Free PMC article.
-
[Microarray-based transcriptome analyses in infectious diseases. A new diagnostic method].Internist (Berl). 2006 Jun;47 Suppl 1:S6, S8-13. doi: 10.1007/s00108-006-1627-6. Internist (Berl). 2006. PMID: 16773368 Review. German.
-
Microarray study reveals that HIV-1 induces rapid type-I interferon-dependent p53 mRNA up-regulation in human primary CD4+ T cells.Retrovirology. 2009 Jan 15;6:5. doi: 10.1186/1742-4690-6-5. Retrovirology. 2009. PMID: 19146679 Free PMC article.
-
Antigen-specific gene expression profiles of peripheral blood mononuclear cells do not reflect those of T-lymphocyte subsets.Clin Diagn Lab Immunol. 2004 Sep;11(5):977-82. doi: 10.1128/CDLI.11.5.977-982.2004. Clin Diagn Lab Immunol. 2004. PMID: 15358662 Free PMC article.
-
Human immunodeficiency virus type-1 Tat protein induces secretory leukocyte protease inhibitor expression in African green monkey but not human cells.Virus Genes. 2020 Apr;56(2):182-193. doi: 10.1007/s11262-020-01731-x. Epub 2020 Jan 10. Virus Genes. 2020. PMID: 31925640
References
-
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. - PubMed
-
- Perou CM, Jeffrey SS, van de Rijn M, Rees CA, Eisen MB, Ross DT, Pergamenschikov A, Williams CF, Zhu SX, Lee JC, Lashkari D, Shalon D, Brown PO, Botstein D. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc Natl Acad Sci U S A. 1999;96:9212–9217. doi: 10.1073/pnas.96.16.9212. - DOI - PMC - PubMed
-
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

