ATP-dependent removal of nucleoside reverse transcriptase inhibitors by human immunodeficiency virus type 1 reverse transcriptase
- PMID: 12069972
- PMCID: PMC127313
- DOI: 10.1128/AAC.46.7.2179-2184.2002
ATP-dependent removal of nucleoside reverse transcriptase inhibitors by human immunodeficiency virus type 1 reverse transcriptase
Abstract
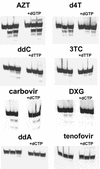
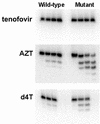
Removal of nucleoside chain terminator inhibitors mediated by human immunodeficiency virus (HIV) reverse transcriptase (RT) using ATP as an acceptor molecule has been proposed as a novel mechanism of HIV resistance. Recombinant wild-type and mutant HIV type 1 (HIV-1) RT enzymes with thymidine analog resistance mutations D67N, K70R, and T215Y were analyzed for their ability to remove eight nucleoside reverse transcriptase inhibitors in the presence of physiological concentrations of ATP. The order for the rate of removal of the eight inhibitors by the mutant RT enzyme was zidovudine (AZT) > stavudine (d4T) >> zalcitabine (ddC) > abacavir > amdoxovir (DAPD) > lamivudine (3TC) > didanosine (ddI) > tenofovir. Thymidine analogs AZT and d4T were the most significantly removed by the mutant enzyme, suggesting that removal of these inhibitors by the ATP-dependent removal mechanism contributes to the AZT and d4T resistance observed in patients with HIV expressing thymidine analog resistance mutations. ATP-dependent removal of tenofovir was 22- to 35-fold less efficient than removal of d4T and AZT, respectively. The addition of ATP and the next complementary deoxynucleoside triphosphate caused a reduction of ATP-mediated removal of d4T, ddC, and DAPD, while AZT and abacavir removal was unaffected. The reduction of d4T, ddC, and DAPD removal in the presence of the deoxynucleoside triphosphate could explain the minor changes in susceptibility to these drugs observed in conventional in vitro phenotypic assays using cells that have higher deoxynucleoside triphosphate pools. The minimal removal of abacavir, ddC, DAPD, 3TC, ddI, and tenofovir is consistent with the minor changes in susceptibility to these drugs observed for HIV mutants with thymidine analog resistance mutations.
Figures


References
-
- Arion, D., N. Kaushik, S. McCormick, G. Borkow, and M. A. Parniak. 1998. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry 37:15908-15917. - PubMed
-
- Barshop, B. A., D. T. Adamson, D. C. Vellom, F. Rosen, B. L. Epstein, and J. E. Seegmiller. 1991. Luminescent immobilized enzyme test systems for inorganic pyrophosphate: assays using firefly luciferase and nicotinamide-mononucleotide adenylyl transferase or adenosine-5′-triphosphate sulfurylase. Anal. Biochem. 197:266-272. - PubMed
-
- Battula, N., and L. A. Loeb. 1976. On the fidelity of DNA replication. Lack of exodeoxyribonuclease activity and error-correcting function in avian myeloblastosis virus DNA polymerase. J. Biol. Chem. 251:982-986. - PubMed
-
- Boucher, C. A. B., W. Keulen, T. van Bommel, M. Nijhuis, D. de Jong, M. D. de Jong, P. Schipper, and N. K. T. Back. 1996. HIV-1 drug susceptibility determination by using recombinant viruses generated from patient sera tested in a cell-killing assay. Antimicrob. Agents Chemother. 40:2404-2409. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

