Diversity of beta-lactam resistance-conferring amino acid substitutions in penicillin-binding protein 3 of Haemophilus influenzae
- PMID: 12069976
- PMCID: PMC127296
- DOI: 10.1128/AAC.46.7.2208-2218.2002
Diversity of beta-lactam resistance-conferring amino acid substitutions in penicillin-binding protein 3 of Haemophilus influenzae
Abstract
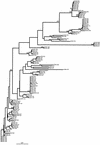
The sequences of the ftsI gene, encoding the transpeptidase domain of penicillin binding protein (PBP) 3A and/or PBP 3B, which are involved in septal peptidoglycan synthesis, were determined for 108 clinical strains of Haemophilus influenzae with reduced susceptibility to beta-lactam antibiotics with or without beta-lactamase production and were compared to those of the ampicillin-susceptible Rd strain and ampicillin-susceptible clinical isolates. The sequences have 18 different mutation patterns and were classified into two groups on the basis of amino acid substitutions deduced from the nucleotide sequences located between bp 960 and 1618 of the ftsI gene. In group I strains (n = 7), His-517 was substituted for Arg-517. In group II strains (n = 101), Lys-526 was substituted for Asn-526. In subgroup IIa (n = 5; H. influenzae ATCC 49247), the only observed substitution was Lys-526 for Asn-526; in subgroup IIb (n = 56), Val-502 was substituted for Ala-502 (n = 13), along with several other substitutions: Asn-350 for Asp-350 (n = 15), Asn-350 for Asp-350 and Glu-490 for Gly-490 (n = 14), and Asn-350 for Asp-350 and Ser-437 for Ala-437 (n = 5). In subgroup IIc (n = 25), Thr-502 was substituted for Ala-502. In subgroup IId, Val-449 was substituted for Ile-449 (n = 15). The MICs of beta-lactam antibiotics for the 108 strains were to 8 to 16 times the MICs for susceptible strains. The strains, isolated from both adults and children, were analyzed for genetic relationship by pulsed-field gel electrophoresis and by determination of ftsI sequence phylogeny. Both analyses revealed the lack of clonality and the heterogeneity of the strains, but some clusters suggest the spread and/or persistence of a limited number of strains of the same pulsotype and pattern of amino acid substitutions. Reduced susceptibility to beta-lactam, brought about by mutations of the ftsI gene, is becoming a frequent phenomenon, affecting both strains that produce beta-lactamase and those that do not. The level of resistance remains low but opens the way to greater resistance in the future.
Figures


References
-
- Bell, S. M., and D. Plowman. 1980. Mechanisms of ampicillin resistance in Haemophilus influenzae from respiratory tract. Lancet i:279-280. - PubMed
-
- Campos, J., F. Román, M. Georgiou, C. Garcia, R. Gómez-Lus, R. Cantón, H. Escobar, and F. Baquero. 1996. Long-term persistence of ciprofloxacin-resistant Haemophilus influenzae in patients with cystic fibrosis. J. Infect. Dis. 174:1345-1347. - PubMed
-
- Clairoux, N., M. Picard, A. Brochu, N. Rousseau, P. Gourde, D. Beauchamp, T. R. Parr, M. G. Bergeron, and F. Malouin. 1992. Molecular basis of the non-β-lactamase-mediated resistance to β-lactam antibiotics in strains of Haemophilus influenzae isolated in Canada. Antimicrob. Agents Chemother. 36:1504-1513. - PMC - PubMed
-
- Dabernat, H., and C. Delmas. 1998. Activité du Centre National de Référence des Haemophilus influenzae, années 1996-1997: le déclin du type b. Med. Mal. Infect. 28:940-946.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

