High-throughput screening of soluble recombinant proteins
- PMID: 12070324
- PMCID: PMC2373646
- DOI: 10.1110/ps.0205202
High-throughput screening of soluble recombinant proteins
Abstract
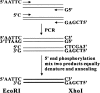
The aims of high-throughput (HTP) protein production systems are to obtain well-expressed and highly soluble proteins, which are preferred candidates for use in structure-function studies. Here, we describe the development of an efficient and inexpensive method for parallel cloning, induction, and cell lysis to produce multiple fusion proteins in Escherichia coli using a 96-well format. Molecular cloning procedures, used in this HTP system, require no restriction digestion of the PCR products. All target genes can be directionally cloned into eight different fusion protein expression vectors using two universal restriction sites and with high efficiency (>95%). To screen for well-expressed soluble fusion protein, total cell lysates of bacteria culture ( approximately 1.5 mL) were subjected to high-speed centrifugation in a 96-tube format and analyzed by multiwell denaturing SDS-PAGE. Our results thus far show that 80% of the genes screened show high levels of expression of soluble products in at least one of the eight fusion protein constructs. The method is well suited for automation and is applicable for the production of large numbers of proteins for genome-wide analysis.
Figures




Similar articles
-
High-throughput protein expression screening and purification in Escherichia coli.Methods. 2011 Sep;55(1):65-72. doi: 10.1016/j.ymeth.2011.08.010. Epub 2011 Sep 8. Methods. 2011. PMID: 21925268
-
Screening for soluble expression of recombinant proteins in a 96-well format.Anal Biochem. 2001 Oct 1;297(1):79-85. doi: 10.1006/abio.2001.5331. Anal Biochem. 2001. PMID: 11567530
-
The pURI family of expression vectors: a versatile set of ligation independent cloning plasmids for producing recombinant His-fusion proteins.Protein Expr Purif. 2011 Mar;76(1):44-53. doi: 10.1016/j.pep.2010.10.013. Epub 2010 Nov 3. Protein Expr Purif. 2011. PMID: 21055470
-
Tuning different expression parameters to achieve soluble recombinant proteins in E. coli: advantages of high-throughput screening.Biotechnol J. 2011 Jun;6(6):715-30. doi: 10.1002/biot.201100025. Epub 2011 May 12. Biotechnol J. 2011. PMID: 21567962 Review.
-
Breaking the bottleneck: eukaryotic membrane protein expression for high-resolution structural studies.J Struct Biol. 2007 Dec;160(3):265-74. doi: 10.1016/j.jsb.2007.07.001. Epub 2007 Jul 14. J Struct Biol. 2007. PMID: 17702603 Review.
Cited by
-
Learning to predict expression efficacy of vectors in recombinant protein production.BMC Bioinformatics. 2010 Jan 18;11 Suppl 1(Suppl 1):S21. doi: 10.1186/1471-2105-11-S1-S21. BMC Bioinformatics. 2010. PMID: 20122193 Free PMC article.
-
Production Optimization of an Active β-Galactosidase of Bifidobacterium animalis in Heterologous Expression Systems.Biomed Res Int. 2019 Feb 20;2019:8010635. doi: 10.1155/2019/8010635. eCollection 2019. Biomed Res Int. 2019. PMID: 30915359 Free PMC article.
-
Self-assembly of MinE on the membrane underlies formation of the MinE ring to sustain function of the Escherichia coli Min system.J Biol Chem. 2014 Aug 1;289(31):21252-66. doi: 10.1074/jbc.M114.571976. Epub 2014 Jun 9. J Biol Chem. 2014. PMID: 24914211 Free PMC article.
-
Engineering a novel endopeptidase based on SARS 3CL(pro).Biotechniques. 2009 Dec;47(6):1029-32. doi: 10.2144/000113303. Biotechniques. 2009. PMID: 20041855 Free PMC article.
-
Enhancement of solubility in Escherichia coli and purification of an aminotransferase from Sphingopyxis sp. MTA144 for deamination of hydrolyzed fumonisin B(1).Microb Cell Fact. 2010 Aug 18;9:62. doi: 10.1186/1475-2859-9-62. Microb Cell Fact. 2010. PMID: 20718948 Free PMC article.
References
-
- Christendat, D., Yee, A., Dharamsi, A., Kluger, Y., Savchenko, A., Cort, J.R., Booth, V., Mackereth, C.D., Saridakis, V., Ekiel, I., et al. 2000. Structural proteomics of an archaeon. Nat. Struct. Biol. 7 903–909. - PubMed
-
- Edwards, A.M. Arrowsmith, C.H. Christendat, D. Dharamsi, A., Friesen, J.D. Greenblatt, J.F., and Vedadi, M. 2000. Protein production: Feeding the crystallographers, and NMR spectroscopies. Nat. Struct. Biol. [Suppl.] 7 970–972. - PubMed
-
- Fields, S. 2001. Proteomics: Proteomics in genomeland. Science 291 1221–1224. - PubMed
-
- Lesley, S.A. 2001. High-throughput proteomics: Protein expression and purification in the postgenomic world. Protein Expr. Purif. 22 159–164. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

