Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis
- PMID: 12070350
- PMCID: PMC124423
- DOI: 10.1073/pnas.132092099
Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis
Abstract
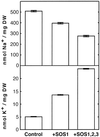
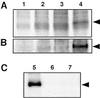
The Arabidopsis thaliana SOS1 protein is a putative Na+/H+ antiporter that functions in Na+ extrusion and is essential for the NaCl tolerance of plants. sos1 mutant plants share phenotypic similarities with mutants lacking the protein kinase SOS2 and the Ca2+ sensor SOS3. To investigate whether the three SOS proteins function in the same response pathway, we have reconstituted the SOS system in yeast cells. Expression of SOS1 improved the Na+ tolerance of yeast mutants lacking endogenous Na+ transporters. Coexpression of SOS2 and SOS3 dramatically increased SOS1-dependent Na+ tolerance, whereas SOS2 or SOS3 individually had no effect. The SOS2/SOS3 kinase complex promoted the phosphorylation of SOS1. A constitutively active form of SOS2 phosphorylated SOS1 in vitro independently of SOS3, but could not fully substitute for the SOS2/SOS3 kinase complex for activation of SOS1 in vivo. Further, we show that SOS3 recruits SOS2 to the plasma membrane. Although sos1 mutant plants display defective K+ uptake at low external concentrations, neither the unmodified nor the SOS2/SOS3-activated SOS1 protein showed K+ transport capacity in vivo, suggesting that the role of SOS1 on K+ uptake is indirect. Our results provide an example of functional reconstitution of a plant response pathway in a heterologous system and demonstrate that the SOS1 ion transporter, the SOS2 protein kinase, and its associated Ca2+ sensor SOS3 constitute a functional module. We propose a model in which SOS3 activates and directs SOS2 to the plasma membrane for the stimulatory phosphorylation of the Na+ transporter SOS1.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

