A comprehensive analysis of recently integrated human Ta L1 elements
- PMID: 12070800
- PMCID: PMC379164
- DOI: 10.1086/341718
A comprehensive analysis of recently integrated human Ta L1 elements
Abstract
The Ta (transcribed, subset a) subfamily of L1 LINEs (long interspersed elements) is characterized by a 3-bp ACA sequence in the 3' untranslated region and contains approximately 520 members in the human genome. Here, we have extracted 468 Ta L1Hs (L1 human specific) elements from the draft human genomic sequence and screened individual elements using polymerase-chain-reaction (PCR) assays to determine their phylogenetic origin and levels of human genomic diversity. One hundred twenty-four of the elements amenable to complete sequence analysis were full length ( approximately 6 kb) and have apparently escaped any 5' truncation. Forty-four of these full-length elements have two intact open reading frames and may be capable of retrotransposition. Sequence analysis of the Ta L1 elements showed a low level of nucleotide divergence with an estimated age of 1.99 million years, suggesting that expansion of the L1 Ta subfamily occurred after the divergence of humans and African apes. A total of 262 Ta L1 elements were screened with PCR-based assays to determine their phylogenetic origin and the level of human genomic variation associated with each element. All of the Ta L1 elements analyzed by PCR were absent from the orthologous positions in nonhuman primate genomes, except for a single element (L1HS72) that was also present in the common (Pan troglodytes) and pygmy (P. paniscus) chimpanzee genomes. Sequence analysis revealed that this single exception is the product of a gene conversion event involving an older preexisting L1 element. One hundred fifteen (45%) of the Ta L1 elements were polymorphic with respect to insertion presence or absence and will serve as identical-by-descent markers for the study of human evolution.
Figures


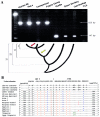

Similar articles
-
LINE-1 preTa elements in the human genome.J Mol Biol. 2003 Feb 28;326(4):1127-46. doi: 10.1016/s0022-2836(03)00032-9. J Mol Biol. 2003. PMID: 12589758
-
Following the LINEs: an analysis of primate genomic variation at human-specific LINE-1 insertion sites.Mol Biol Evol. 2003 Aug;20(8):1338-48. doi: 10.1093/molbev/msg146. Epub 2003 May 30. Mol Biol Evol. 2003. PMID: 12777507
-
Reading between the LINEs: human genomic variation induced by LINE-1 retrotransposition.Genome Res. 2000 Oct;10(10):1496-508. doi: 10.1101/gr.149400. Genome Res. 2000. PMID: 11042149 Free PMC article.
-
Alu repeats and human genomic diversity.Nat Rev Genet. 2002 May;3(5):370-9. doi: 10.1038/nrg798. Nat Rev Genet. 2002. PMID: 11988762 Review.
-
A systematic analysis of LINE-1 endonuclease-dependent retrotranspositional events causing human genetic disease.Hum Genet. 2005 Sep;117(5):411-27. doi: 10.1007/s00439-005-1321-0. Epub 2005 Jun 28. Hum Genet. 2005. PMID: 15983781 Review.
Cited by
-
The NF1 gene contains hotspots for L1 endonuclease-dependent de novo insertion.PLoS Genet. 2011 Nov;7(11):e1002371. doi: 10.1371/journal.pgen.1002371. Epub 2011 Nov 17. PLoS Genet. 2011. PMID: 22125493 Free PMC article.
-
Mammalian small nucleolar RNAs are mobile genetic elements.PLoS Genet. 2006 Dec 8;2(12):e205. doi: 10.1371/journal.pgen.0020205. Epub 2006 Oct 20. PLoS Genet. 2006. PMID: 17154719 Free PMC article.
-
Genomic rearrangements by LINE-1 insertion-mediated deletion in the human and chimpanzee lineages.Nucleic Acids Res. 2005 Jul 20;33(13):4040-52. doi: 10.1093/nar/gki718. Print 2005. Nucleic Acids Res. 2005. PMID: 16034026 Free PMC article.
-
Tangram: a comprehensive toolbox for mobile element insertion detection.BMC Genomics. 2014 Sep 16;15(1):795. doi: 10.1186/1471-2164-15-795. BMC Genomics. 2014. PMID: 25228379 Free PMC article.
-
The genomic distribution of L1 elements: the role of insertion bias and natural selection.J Biomed Biotechnol. 2006;2006(1):75327. doi: 10.1155/JBB/2006/75327. J Biomed Biotechnol. 2006. PMID: 16877820 Free PMC article.
References
Electronic-Database Information
-
- Batzer Lab, http://batzerlab.lsu.edu/
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for the DNA sequences from the common and pygmy chimpanzee orthologs of L1HS72 [accession numbers AF489459 and AF489460]; diverse DNA sequences from L1HS72 [accession numbers AF489450–AF489458]; and Ta L1 element pre-integration site sequences, namely, L1HS45 [accession numbers AF461364 and AF461365], L1HS172 [accession numbers AF461368 and AF461369], L1HS178 [accession numbers AF461370 and AF461371], L1HS284 [accession numbers AF461372 and AF461373], L1HS372 [accession numbers AF461374 and AF461375], L1HS416 [accession numbers AF461376 and AF461377], L1HS442 [accession numbers AF461378 and AF461379], L1HS443 [accession numbers AF461386 and AF461387], L1HS513 [accession numbers AF461380–AF461382], and L1HS558 [accession number AF461383])
-
- Genetic Information Research Institute Censor Server, http://www.girinst.org/Censor_Server-Data_Entry_Forms.html
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410 - PubMed
-
- Arcot SS, Wang Z, Weber JL, Deininger PL, Batzer MA (1995) Alu repeats: a source for the genesis of primate microsatellites. Genomics 29:136–144 - PubMed
-
- Ausabel FM, Brent R, Kingston ME, Moore DD, Seidman JG (1987) Current protocols in molecular biology. John Wiley & Sons, New York
-
- Batzer MA, Deininger PL (2002) Alu repeats and human genomic diversity. Nat Rev Genet 3:370–379 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

