Template requirements for de novo RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase on the viral X RNA
- PMID: 12072495
- PMCID: PMC136341
- DOI: 10.1128/jvi.76.14.6944-6956.2002
Template requirements for de novo RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase on the viral X RNA
Abstract
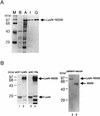
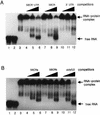
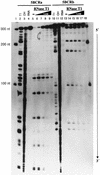
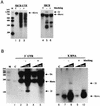
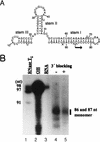
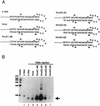
The hepatitis C virus (HCV)-encoded NS5B protein is an RNA-dependent RNA polymerase which plays a substantial role in viral replication. We expressed and purified the recombinant NS5B of an HCV genotype 3a from Esherichia coli, and we investigated its ability to bind to the viral RNA and its enzymatic activity. The results presented here demonstrate that NS5B interacts strongly with the coding region of positive-strand RNA, although not in a sequence-specific manner. It was also determined that more than two molecules of polymerase bound sequentially to this region with the direction 3' to 5'. Also, we attempted to determine the initiation site(s) of de novo synthesis by NS5B on X RNA, which contains the last 98 nucleotides of HCV positive-strand RNA. The initiation site(s) on X RNA was localized in the pyrimidine-rich region of stem I. However, when more than five of the nucleotides of stem I in X RNA were deleted from the 3' end, RNA synthesis initiated at another site of the specific ribonucleotide. Our study also showed that the efficiency of RNA synthesis, which was directed by X RNA, was maximized by the GC base pair at the penultimate position from the 3' end of the stem. These results will provide some clues to understanding the mechanism of HCV genomic RNA replication in terms of viral RNA-NS5B interaction and the initiation of de novo RNA synthesis.
Figures




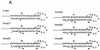
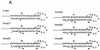
References
-
- Arnold, J. J., and C. E. Cameron. 1999. Poliovirus RNA-dependent RNA polymerase (3Dpol) is sufficient for template switching in vitro. J. Biol. Chem. 274:2706-2716. - PubMed
-
- Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

