Selection of 3'-template bases and initiating nucleotides by hepatitis C virus NS5B RNA-dependent RNA polymerase
- PMID: 12072503
- PMCID: PMC136306
- DOI: 10.1128/jvi.76.14.7030-7039.2002
Selection of 3'-template bases and initiating nucleotides by hepatitis C virus NS5B RNA-dependent RNA polymerase
Abstract
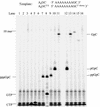
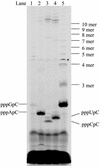
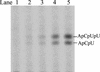
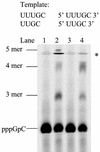
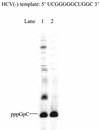
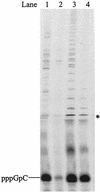
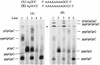
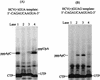
De novo RNA synthesis by hepatitis C virus (HCV) nonstructural protein 5B (NS5B) RNA-dependent RNA polymerase has been investigated using short RNA templates. Various templates including those derived from the HCV genome were evaluated by examining the early steps of de novo RNA synthesis. NS5B was shown to be able to produce an initiation dinucleotide product from templates as short as 4-mer and from the 3'-terminal sequences of both plus and minus strands of the HCV RNA genome. GMP, GDP, and guanosine were able to act as an initiating nucleotide in de novo RNA synthesis, indicating that the triphosphate moiety is not absolutely required by an initiating nucleotide. Significant amounts of the initiation product accumulated in de novo synthesis, and elongation from the dinucleotide was observed when large amounts of dinucleotide were available. This result suggests that NS5B, a template, and incoming nucleotides are able to form an initiation complex that aborts frequently by releasing the dinucleotide product before transition to an elongation complex. The transition is rate limiting. Furthermore, we discovered that the secondary structure of a template was not essential for de novo initiation and that 3'-terminal bases of a template conferred specificity in selection of an initiation site. Initiation can occur at the +1, +2, or +3 position numbered from the 3' end of a template depending on base composition. Pyrimidine bases at any of the three positions are able to serve as an initiation site, while purine bases at the +2 and +3 positions do not support initiation. This result implies that HCV possesses an intrinsic ability to ensure that de novo synthesis is initiated from the +1 position and to maintain the integrity of the 3' end of its genome. This assay system should be an important tool for investigating the detailed mechanism of de novo initiation by HCV NS5B as well as other viral RNA polymerases.
Figures


Similar articles
-
Template requirements for de novo RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase on the viral X RNA.J Virol. 2002 Jul;76(14):6944-56. doi: 10.1128/jvi.76.14.6944-6956.2002. J Virol. 2002. PMID: 12072495 Free PMC article.
-
Template requirement and initiation site selection by hepatitis C virus polymerase on a minimal viral RNA template.J Biol Chem. 2000 Jun 9;275(23):17710-7. doi: 10.1074/jbc.M908781199. J Biol Chem. 2000. PMID: 10749880
-
De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus.J Virol. 2000 Jan;74(2):851-63. doi: 10.1128/jvi.74.2.851-863.2000. J Virol. 2000. PMID: 10623748 Free PMC article.
-
Hepatitis C virus RNA-dependent RNA polymerase (NS5B polymerase).Curr Top Microbiol Immunol. 2000;242:225-60. doi: 10.1007/978-3-642-59605-6_11. Curr Top Microbiol Immunol. 2000. PMID: 10592663 Review. No abstract available.
-
Biochemical and structural analysis of the NS5B RNA-dependent RNA polymerase of the hepatitis C virus.J Viral Hepat. 2000 May;7(3):167-74. doi: 10.1046/j.1365-2893.2000.00218.x. J Viral Hepat. 2000. PMID: 10849258 Review.
Cited by
-
Form confers function: Case of the 3'X region of the hepatitis C virus genome.World J Gastroenterol. 2018 Aug 14;24(30):3374-3383. doi: 10.3748/wjg.v24.i30.3374. World J Gastroenterol. 2018. PMID: 30122877 Free PMC article. Review.
-
Structure and function of the 3' terminal six nucleotides of the west nile virus genome in viral replication.J Virol. 2004 Aug;78(15):8159-71. doi: 10.1128/JVI.78.15.8159-8171.2004. J Virol. 2004. PMID: 15254187 Free PMC article.
-
Structural explanation for the role of Mn2+ in the activity of phi6 RNA-dependent RNA polymerase.Nucleic Acids Res. 2008 Nov;36(20):6633-44. doi: 10.1093/nar/gkn632. Epub 2008 Oct 21. Nucleic Acids Res. 2008. PMID: 18940872 Free PMC article.
-
Stimulation of interferon-β responses by aberrant SARS-CoV-2 small viral RNAs acting as retinoic acid-inducible gene-I agonists.iScience. 2023 Jan 20;26(1):105742. doi: 10.1016/j.isci.2022.105742. Epub 2022 Dec 7. iScience. 2023. PMID: 36507221 Free PMC article.
-
Factors affecting de novo RNA synthesis and back-priming by the respiratory syncytial virus polymerase.Virology. 2014 Aug;462-463:318-27. doi: 10.1016/j.virol.2014.05.032. Epub 2014 Jul 8. Virology. 2014. PMID: 25010481 Free PMC article.
References
-
- Ago, H., T. Adachi, A. Yoshida, M. Yamamoto, N. Habuka, K. Yatsunami, and M. Miyano. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Struct. Fold Des. 7:1417-1426. - PubMed
-
- Butcher, S. J., J. M. Grimes, E. V. Makeyev, D. H. Bamford, and D. I. Stuart. 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410:235-240. - PubMed
-
- Hong, Z., C. E. Cameron, M. P. Walker, C. Castro, N. Yao, J. Y. Lau, and W. Zhong. 2001. A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology 285:6-11. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

