Processing map and essential cleavage sites of the nonstructural polyprotein encoded by ORF1 of the feline calicivirus genome
- PMID: 12072506
- PMCID: PMC136307
- DOI: 10.1128/jvi.76.14.7060-7072.2002
Processing map and essential cleavage sites of the nonstructural polyprotein encoded by ORF1 of the feline calicivirus genome
Abstract
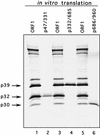
Feline calicivirus (FCV) nonstructural proteins are translated as part of a large polyprotein that undergoes autocatalytic processing by the virus-encoded 3C-like proteinase. In this study, we mapped three new cleavage sites (E(46)/A(47), E(331)/D(332), and E(685)/N(686)) recognized by the virus proteinase in the N-terminal part of the open reading frame 1 (ORF1) polyprotein to complete the processing map. Taken together with two sites we identified previously (E(960)/A(961) and E(1071)/S(1072)), the FCV ORF1 polyprotein contains five cleavage sites that define the borders of six proteins with calculated molecular masses of 5.6, 32, 38.9, 30.1, 12.7, and 75.7 kDa, which we designated p5.6, p32, p39 (NTPase), p30, p13 (VPg), and p76 (Pro-Pol), respectively. Mutagenesis of the E to A in each of these cleavage sites in an infectious FCV cDNA clone was lethal for the virus, indicating that these cleavages are essential in a productive virus infection. Mutagenesis of two cleavage sites (E(1345)/T(1346) and E(1419)/G(1420)) within the 75.7-kDa Pro-Pol protein previously mapped in bacterial expression studies was not lethal.
Figures









Similar articles
-
In vitro proteolytic processing of the MD145 norovirus ORF1 nonstructural polyprotein yields stable precursors and products similar to those detected in calicivirus-infected cells.J Virol. 2003 Oct;77(20):10957-74. doi: 10.1128/jvi.77.20.10957-10974.2003. J Virol. 2003. PMID: 14512545 Free PMC article.
-
Mapping of the feline calicivirus proteinase responsible for autocatalytic processing of the nonstructural polyprotein and identification of a stable proteinase-polymerase precursor protein.J Virol. 1999 Aug;73(8):6626-33. doi: 10.1128/JVI.73.8.6626-6633.1999. J Virol. 1999. PMID: 10400760 Free PMC article.
-
Cleavage map and proteolytic processing of the murine norovirus nonstructural polyprotein in infected cells.J Virol. 2006 Aug;80(16):7816-31. doi: 10.1128/JVI.00532-06. J Virol. 2006. PMID: 16873239 Free PMC article.
-
Proteolytic processing of sapovirus ORF1 polyprotein.J Virol. 2005 Jun;79(12):7283-90. doi: 10.1128/JVI.79.12.7283-7290.2005. J Virol. 2005. PMID: 15919882 Free PMC article.
-
Identification of the 3C-protease-mediated 2A/2B and 2B/2C cleavage sites in the nonstructural polyprotein precursor of a Dicistrovirus lacking the NPGP motif.Arch Virol. 2010 Sep;155(9):1477-82. doi: 10.1007/s00705-010-0723-z. Epub 2010 Jun 13. Arch Virol. 2010. PMID: 20549262
Cited by
-
In vitro proteolytic processing of the MD145 norovirus ORF1 nonstructural polyprotein yields stable precursors and products similar to those detected in calicivirus-infected cells.J Virol. 2003 Oct;77(20):10957-74. doi: 10.1128/jvi.77.20.10957-10974.2003. J Virol. 2003. PMID: 14512545 Free PMC article.
-
Characterization of a strain of feline calicivirus isolated from a dog faecal sample.Vet Microbiol. 2009 Oct 20;139(1-2):52-7. doi: 10.1016/j.vetmic.2009.04.033. Epub 2009 May 5. Vet Microbiol. 2009. PMID: 19481376 Free PMC article.
-
GII.4 Norovirus Protease Shows pH-Sensitive Proteolysis with a Unique Arg-His Pairing in the Catalytic Site.J Virol. 2019 Mar 5;93(6):e01479-18. doi: 10.1128/JVI.01479-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30626675 Free PMC article.
-
Functional analysis of the 5' genomic sequence of a bovine norovirus.PLoS One. 2008 May 14;3(5):e2169. doi: 10.1371/journal.pone.0002169. PLoS One. 2008. PMID: 18478070 Free PMC article.
-
The Norovirus NS3 Protein Is a Dynamic Lipid- and Microtubule-Associated Protein Involved in Viral RNA Replication.J Virol. 2017 Jan 18;91(3):e02138-16. doi: 10.1128/JVI.02138-16. Print 2017 Feb 1. J Virol. 2017. PMID: 27881660 Free PMC article.
References
-
- Aldabe, R., A. Barco, and L. Carrasco. 1996. Membrane permeabilization by poliovirus proteins 2B and 2BC. J. Biol. Chem. 271:23134-23137. - PubMed
-
- Cho, M. W., N. Teterina, D. Egger, K. Bienz, and E. Ehrenfeld. 1994. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology 202:129-145. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

