The RNA polymerase of influenza a virus is stabilized by interaction with its viral RNA promoter
- PMID: 12072510
- PMCID: PMC136304
- DOI: 10.1128/jvi.76.14.7103-7113.2002
The RNA polymerase of influenza a virus is stabilized by interaction with its viral RNA promoter
Abstract
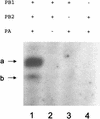
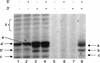
The RNA polymerase of the influenza virus is responsible for the transcription and replication of the segmented RNA viral genome during infection of host cells. Polymerase function is known to be strictly dependent on interaction with its RNA promoter, but no attempts to investigate whether the virion RNA (vRNA) promoter stabilizes the polymerase have been reported previously. Here we tested whether the vRNA promoter protects the polymerase against heat inactivation. We prepared partially purified recombinant influenza A virus RNA polymerase, in the absence of influenza virus vRNA promoter sequences, by transient transfection of expression plasmids into human kidney 293T cells. The polymerase was found to be heat labile at 40 degrees C in the absence of added vRNA. However, it was protected from heat inactivation if both the 5' and 3' strands of the vRNA promoter were present. By using the ability of vRNA to protect the enzyme against heat inactivation, we established a novel assay, in conjunction with a mutagenic approach, that was used to test the secondary structure requirement of the vRNA promoter for polymerase binding. Binding required a panhandle structure and the presence of local hairpin loop structures in both the 5' and 3' ends of vRNA, as suggested by the corkscrew model. The interaction of the vRNA promoter with the influenza virus RNA polymerase heterotrimeric complex is likely to favor a particular closed conformation of the complex, thereby ensuring the stability of the RNA polymerase within both the infected cell and the isolated virus.
Figures




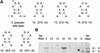

Similar articles
-
Solution structure of the influenza A virus cRNA promoter: implications for differential recognition of viral promoter structures by RNA-dependent RNA polymerase.Nucleic Acids Res. 2003 Jun 1;31(11):2824-32. doi: 10.1093/nar/gkg387. Nucleic Acids Res. 2003. PMID: 12771209 Free PMC article.
-
Interaction of influenza virus polymerase with viral RNA in the 'corkscrew' conformation.J Gen Virol. 1999 Oct;80 ( Pt 10):2565-2572. doi: 10.1099/0022-1317-80-10-2565. J Gen Virol. 1999. PMID: 10573148
-
Temperature sensitive influenza A virus genome replication results from low thermal stability of polymerase-cRNA complexes.Virol J. 2006 Aug 25;3:58. doi: 10.1186/1743-422X-3-58. Virol J. 2006. PMID: 16934156 Free PMC article.
-
Structural insights into RNA synthesis by the influenza virus transcription-replication machine.Virus Res. 2017 Apr 15;234:103-117. doi: 10.1016/j.virusres.2017.01.013. Epub 2017 Jan 20. Virus Res. 2017. PMID: 28115197 Review.
-
Transcription and replication of the influenza a virus genome.Acta Virol. 2000 Oct;44(5):273-82. Acta Virol. 2000. PMID: 11252672 Review.
Cited by
-
The influenza A segment 7 mRNA 3' splice site pseudoknot/hairpin family.RNA Biol. 2012 Nov;9(11):1305-10. doi: 10.4161/rna.22343. Epub 2012 Oct 12. RNA Biol. 2012. PMID: 23064116 Free PMC article.
-
Impact of the segment-specific region of the 3'-untranslated region of the influenza A virus PB1 segment on protein expression.Virus Genes. 2013 Dec;47(3):429-38. doi: 10.1007/s11262-013-0969-0. Epub 2013 Aug 15. Virus Genes. 2013. PMID: 23949786
-
The RNA-dependent RNA polymerase of the influenza A virus.Future Virol. 2014 Sep;9(9):863-876. doi: 10.2217/fvl.14.66. Future Virol. 2014. PMID: 25431616 Free PMC article.
-
The PA subunit is required for efficient nuclear accumulation of the PB1 subunit of the influenza A virus RNA polymerase complex.J Virol. 2004 Sep;78(17):9144-53. doi: 10.1128/JVI.78.17.9144-9153.2004. J Virol. 2004. PMID: 15308710 Free PMC article.
-
Identification of a PA-binding peptide with inhibitory activity against influenza A and B virus replication.PLoS One. 2009 Oct 20;4(10):e7517. doi: 10.1371/journal.pone.0007517. PLoS One. 2009. PMID: 19841738 Free PMC article.
References
-
- De la Luna, S., J. Martin, A. Portela, and J. Ortin. 1993. Influenza virus naked RNA can be expressed upon transfection into cells co-expressing the three subunits of the polymerase and the nucleoprotein from SV40 recombinant viruses. J. Gen. Virol. 74:535-539. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

