Evidence for segment-nonspecific packaging of the influenza a virus genome
- PMID: 12072513
- PMCID: PMC136309
- DOI: 10.1128/jvi.76.14.7133-7139.2002
Evidence for segment-nonspecific packaging of the influenza a virus genome
Abstract
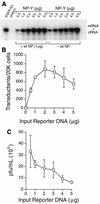
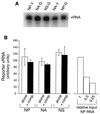
The influenza A virus genome is composed of eight negative-sense RNA segments (called vRNAs), all of which must be packaged to produce an infectious virion. It is not clear whether individual vRNAs are packaged specifically or at random, however, and the total vRNA capacity of the virion is unknown. We have created modified forms of the viral nucleoprotein (NP), neuraminidase (NA), and nonstructural (NS) vRNAs that encode green or yellow fluorescent proteins and studied the efficiency with which these are packaged by using a plasmid-based influenza A virus assembly system. Packaging was assessed precisely and quantitatively by scoring transduction of the fluorescent markers in a single-round infectivity assay with a flow cytometer. We found that, under conditions in which virions are limiting, pairs of alternatively tagged vRNAs compete for packaging but do so in a nonspecific manner. Reporters representing different vRNAs were not packaged additively, as would be expected under specific packaging, but instead appeared to compete for a common niche in the virion. Moreover, 3 to 5% of transduction-competent viruses were found to incorporate two alternative reporters, regardless of whether those reporters represented the same or different vRNAs - a finding compatible with random, but not with specific, packaging. Probabilistic estimates suggest that in order to achieve this level of dual transduction by chance alone, each influenza A virus virion must package an average of 9 to 11 vRNAs.
Figures


References
-
- Chen, W., P. A. Calvo, D. Malide, J. Gibbs, U. Schubert, I. Bacik, S. Basta, R. O'Neill, J. Schickli, P. Palese, P. Henklein, J. R. Bennink, and J. W. Yewdell. 2001. A novel influenza A virus mitochondrial protein that induces cell death. Nat. Med. 7:1306-1312. - PubMed
-
- Compans, R. W., and P. W. Choppin. 1975. Reproduction of myxoviruses, p. 179-252. In H. Fraenkel-Conrat and R. R. Wagner (ed.), Comprehensive virology, vol. 6. Plenum Press, New York, N.Y.
-
- Compans, R. W., N. J. Dimmock, and H. Meier-Ewert. 1970. An electron microscopic study of the influenza virus-infected cell, p. 87-109. In R.D. Barry and B. W. J. Mahy (ed.), The biology of large RNA viruses. Academic Press, London, England.
-
- de la Luna, S., J. Martin, A. Portela, and J. Ortin. 1993. Influenza virus naked RNA can be expressed upon transfection into cells coexpressing the three subunits of the polymerase and the nucleoprotein from simian virus 40 recombinant viruses. J. Gen. Virol. 74:535-539. - PubMed
-
- Duhaut, S., and N. J. Dimmock. 2000. Approximately 150 nucleotides from the 5′ end of an influenza A virus segment 1 defective virion RNA are needed for genome stability during passage of defective virus in infected cells. Virology 275:278-285. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

