Aura virus structure suggests that the T=4 organization is a fundamental property of viral structural proteins
- PMID: 12072523
- PMCID: PMC136343
- DOI: 10.1128/jvi.76.14.7239-7246.2002
Aura virus structure suggests that the T=4 organization is a fundamental property of viral structural proteins
Abstract
Aura and Sindbis viruses are closely related alphaviruses. Unlike other alphaviruses, Aura virus efficiently encapsidates both genomic RNA (11.8 kb) and subgenomic RNA (4.2 kb) to form virus particles. Previous studies on negatively stained Aura virus particles predicted that there were two major size classes with potential T=3 and T=4 capsid structures. We have used cryoelectron microscopy and three-dimensional image reconstruction techniques to examine the native morphology of different classes of Aura virus particles produced in BHK cells. Purified particles separated into two components in a sucrose gradient. Reconstructions of particles in the top and bottom components were computed to resolutions of 17 and 21 A, respectively, and compared with reconstructions of Sindbis virus and Ross River virus particles. Aura virus particles of both top and bottom components have similar, T=4 structures that resemble those of other alphaviruses. The morphology of Aura virus glycoprotein spikes closely resembles that of Sindbis virus spikes and is detectably different from that of Ross River virus spikes. Thus, some aspects of the surface structure of members of the Sindbis virus lineage have been conserved, but other aspects have diverged from the Semliki Forest/Ross River virus lineage.
Figures


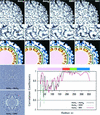
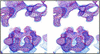
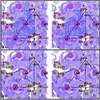

Similar articles
-
A structural and functional perspective of alphavirus replication and assembly.Future Microbiol. 2009 Sep;4(7):837-56. doi: 10.2217/fmb.09.59. Future Microbiol. 2009. PMID: 19722838 Free PMC article. Review.
-
Subgenomic mRNA of Aura alphavirus is packaged into virions.J Virol. 1994 Jan;68(1):56-62. doi: 10.1128/JVI.68.1.56-62.1994. J Virol. 1994. PMID: 7902874 Free PMC article.
-
Aura alphavirus subgenomic RNA is packaged into virions of two sizes.J Virol. 1995 Mar;69(3):1741-6. doi: 10.1128/JVI.69.3.1741-1746.1995. J Virol. 1995. PMID: 7853512 Free PMC article.
-
Complete sequence of the genomic RNA of O'nyong-nyong virus and its use in the construction of alphavirus phylogenetic trees.Virology. 1990 Mar;175(1):110-23. doi: 10.1016/0042-6822(90)90191-s. Virology. 1990. PMID: 2155505
-
The regulation of disassembly of alphavirus cores.Arch Virol. 2009;154(3):381-90. doi: 10.1007/s00705-009-0333-9. Epub 2009 Feb 19. Arch Virol. 2009. PMID: 19225713 Review.
Cited by
-
Alphavirus Particles Can Assemble with an Alternate Triangulation Number.Viruses. 2022 Nov 27;14(12):2650. doi: 10.3390/v14122650. Viruses. 2022. PMID: 36560655 Free PMC article.
-
Determining functionally important amino acid residues of the E1 protein of Venezuelan equine encephalitis virus.J Mol Model. 2006 Sep;12(6):921-9. doi: 10.1007/s00894-006-0101-7. Epub 2006 Apr 11. J Mol Model. 2006. PMID: 16607494
-
A structural and functional perspective of alphavirus replication and assembly.Future Microbiol. 2009 Sep;4(7):837-56. doi: 10.2217/fmb.09.59. Future Microbiol. 2009. PMID: 19722838 Free PMC article. Review.
-
Implication for alphavirus host-cell entry and assembly indicated by a 3.5Å resolution cryo-EM structure.Nat Commun. 2018 Dec 14;9(1):5326. doi: 10.1038/s41467-018-07704-x. Nat Commun. 2018. PMID: 30552337 Free PMC article.
-
Heparin binding sites on Ross River virus revealed by electron cryo-microscopy.Virology. 2005 Feb 20;332(2):511-8. doi: 10.1016/j.virol.2004.11.043. Virology. 2005. PMID: 15680416 Free PMC article.
References
-
- Baker, T. S., and R. H. Cheng. 1996. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 116:120-130. - PubMed
-
- Belnap, D. M., N. H. Olson, and T. S. Baker. 1997. A method for establishing the handedness of biological macromolecules. J. Struct. Biol. 120:44-51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

