Amino acid substitutions in the V domain of nectin-1 (HveC) that impair entry activity for herpes simplex virus types 1 and 2 but not for Pseudorabies virus or bovine herpesvirus 1
- PMID: 12072525
- PMCID: PMC136344
- DOI: 10.1128/jvi.76.14.7255-7262.2002
Amino acid substitutions in the V domain of nectin-1 (HveC) that impair entry activity for herpes simplex virus types 1 and 2 but not for Pseudorabies virus or bovine herpesvirus 1
Abstract
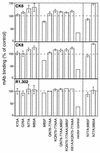
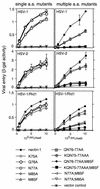
The entry of herpes simplex virus (HSV) into cells requires the interaction of viral glycoprotein D (gD) with a cellular gD receptor to trigger the fusion of viral and cellular membranes. Nectin-1, a member of the immunoglobulin superfamily, can serve as a gD receptor for HSV types 1 and 2 (HSV-1 and HSV-2, respectively) as well as for the animal herpesviruses porcine pseudorabies virus (PRV) and bovine herpesvirus 1 (BHV-1). The HSV-1 gD binding domain of nectin-1 is hypothesized to overlap amino acids 64 to 104 of the N-terminal variable domain-like immunoglobulin domain. Moreover, the HSV-1 and PRV gDs compete for binding to nectin-1. Here we report that two amino acids within this region, at positions 77 and 85, are critical for HSV-1 and HSV-2 entry but not for the entry of PRV or BHV-1. Replacement of either amino acid 77 or amino acid 85 reduced HSV-1 and HSV-2 gD binding but had a lesser effect on HSV entry activity, suggesting that weak interactions between gD and nectin-1 are sufficient to trigger the mechanism of HSV entry. Substitution of both amino acid 77 and amino acid 85 in nectin-1 significantly impaired entry activity for HSV-1 and HSV-2 and eliminated binding to soluble forms of HSV-1 and HSV-2 gDs but did not impair the entry of PRV and BHV-1. Thus, amino acids 77 and 85 of nectin-1 form part of the interface with HSV gD or influence the conformation of that interface. Moreover, the binding sites for HSV and PRV or BHV-1 gDs on nectin-1 may overlap but are not identical.
Figures


Similar articles
-
Glycoprotein D homologs in herpes simplex virus type 1, pseudorabies virus, and bovine herpes virus type 1 bind directly to human HveC(nectin-1) with different affinities.Virology. 2001 Feb 1;280(1):7-18. doi: 10.1006/viro.2000.0747. Virology. 2001. PMID: 11162814
-
Use of chimeric nectin-1(HveC)-related receptors to demonstrate that ability to bind alphaherpesvirus gD is not necessarily sufficient for viral entry.Virology. 2001 Jul 5;285(2):366-75. doi: 10.1006/viro.2001.0989. Virology. 2001. PMID: 11437670
-
Mutations in the N-terminal domains of nectin-1 and nectin-2 reveal differences in requirements for entry of various alphaherpesviruses and for nectin-nectin interactions.J Virol. 2002 Dec;76(24):12940-50. doi: 10.1128/jvi.76.24.12940-12950.2002. J Virol. 2002. PMID: 12438620 Free PMC article.
-
The novel receptors that mediate the entry of herpes simplex viruses and animal alphaherpesviruses into cells.Rev Med Virol. 2000 Sep-Oct;10(5):305-19. doi: 10.1002/1099-1654(200009/10)10:5<305::aid-rmv286>3.0.co;2-t. Rev Med Virol. 2000. PMID: 11015742 Review.
-
Nectin family of cell-adhesion molecules: structural and molecular aspects of function and specificity.Cell Mol Life Sci. 2015 Feb;72(4):645-58. doi: 10.1007/s00018-014-1763-4. Epub 2014 Oct 19. Cell Mol Life Sci. 2015. PMID: 25326769 Free PMC article. Review.
Cited by
-
Role for 3-O-sulfated heparan sulfate as the receptor for herpes simplex virus type 1 entry into primary human corneal fibroblasts.J Virol. 2006 Sep;80(18):8970-80. doi: 10.1128/JVI.00296-06. J Virol. 2006. PMID: 16940509 Free PMC article.
-
Heparanase 2 Modulation Inhibits HSV-2 Replication by Regulating Heparan Sulfate.Viruses. 2024 Nov 26;16(12):1832. doi: 10.3390/v16121832. Viruses. 2024. PMID: 39772142 Free PMC article.
-
Prophylactic, therapeutic and neutralizing effects of zinc oxide tetrapod structures against herpes simplex virus type-2 infection.Antiviral Res. 2012 Dec;96(3):363-75. doi: 10.1016/j.antiviral.2012.09.020. Epub 2012 Oct 6. Antiviral Res. 2012. PMID: 23047013 Free PMC article.
-
In vitro antiviral activity of neem (Azardirachta indica L.) bark extract against herpes simplex virus type-1 infection.Phytother Res. 2010 Aug;24(8):1132-40. doi: 10.1002/ptr.3085. Phytother Res. 2010. PMID: 20041417 Free PMC article.
-
The Ig-like v-type domain of paired Ig-like type 2 receptor alpha is critical for herpes simplex virus type 1-mediated membrane fusion.J Virol. 2010 Sep;84(17):8664-72. doi: 10.1128/JVI.01039-10. Epub 2010 Jun 23. J Virol. 2010. PMID: 20573830 Free PMC article.
References
-
- Arthos, J., K. C. Deen, M. A. Chaikin, J. A. Fornwald, G. Sathe, Q. J. Sattentau, P. R. Clapham, R. A. Weiss, J. S. McDougal, C. Pietropaolo, R. Axel, A. Truneh, P. J. Maddon, and S. R. W. 1989. Identification of the residues in human CD4 critical for the binding of HIV. Cell 57:469-481. - PubMed
-
- Babic, N., B. G. Klupp, B. Makoschey, A. Karger, A. Flamand, and T. C. Mettenleiter. 1996. Glycoprotein gH of pseudorabies virus is essential for penetration and propagation in cell culture and in the nervous system of mice. J. Gen. Virol. 77:2277-2285. - PubMed
-
- Bernhardt, G., J. Harber, A. Zibert, M. deCrombrugghe, and E. Wimmer. 1994. The poliovirus receptor: identification of domains and amino acid residues critical for virus binding. Virology 203:344-356. - PubMed
-
- Campadelli-Fiume, G., F. Cocchi, L. Menotti, and M. Lopez. 2000. The novel receptors that mediate the entry of herpes simplex viruses and animal alphaherpesviruses into cells. Rev. Med. Virol. 10:305-319. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

