Polynucleotide phosphorylase is a global regulator of virulence and persistency in Salmonella enterica
- PMID: 12072563
- PMCID: PMC124376
- DOI: 10.1073/pnas.132047099
Polynucleotide phosphorylase is a global regulator of virulence and persistency in Salmonella enterica
Abstract
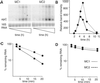
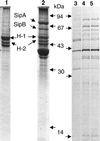
For many pathogens, the ability to regulate their replication in host cells is a key element in establishing persistency. Here, we identified a single point mutation in the gene for polynucleotide phosphorylase (PNPase) as a factor affecting bacterial invasion and intracellular replication, and which determines the alternation between acute or persistent infection in a mouse model for Salmonella enterica infection. In parallel, with microarray analysis, PNPase was found to affect the mRNA levels of a subset of virulence genes, in particular those contained in Salmonella pathogenicity islands 1 and 2. The results demonstrate a connection between PNPase and Salmonella virulence and show that alterations in PNPase activity could represent a strategy for the establishment of persistency.
Figures


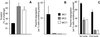
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

