Zinc finger as distance determinant in the flexible linker of intron endonuclease I-TevI
- PMID: 12077294
- PMCID: PMC124309
- DOI: 10.1073/pnas.082253699
Zinc finger as distance determinant in the flexible linker of intron endonuclease I-TevI
Abstract
I-TevI, the phage T4 td intron-encoded endonuclease, recognizes a lengthy DNA target and initiates intron mobility by introducing a double-strand break in the homing site. The enzyme uses both sequence and distance determinants to cleave the DNA 23-25 bp upstream of the intron insertion site. I-TevI consists of an N-terminal catalytic domain and a C-terminal DNA-binding domain separated by a long, flexible linker. The DNA-binding domain consists of three subdomains: a zinc finger, a minor-groove binding alpha-helix, and a helix-turn-helix. In this study, a mutational analysis was undertaken to assess the roles of these subdomains in substrate binding and cleavage. Surprisingly, the zinc finger is not required for DNA binding or catalysis. Rather, the zinc finger is a component of the linker and directs the catalytic domain to cleave the homing site at a fixed distance from the intron insertion site. When the cleavage site (CS) is shifted outside a given range, wild-type I-TevI defaults to the fixed distance, whereas zinc-finger mutants have lost the distance determinant and search out the displaced cleavage sequences. Although counterintuitive, a protein containing a 19-aa deletion of the zinc finger can extend further than can wild-type I-TevI to cleave a distant CS sequence, and a Cys-to-Ala mutant of the ligands for zinc, nominally a longer protein, can retract to cleave at a closer CS sequence. Models are presented for the novel function of the zinc finger, as a molecular constraint, whereby intramolecular protein-protein interactions position the catalytic domain by "catalytic clamp" and/or "linker-organizer" mechanisms.
Figures

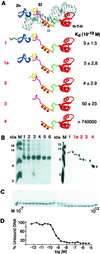
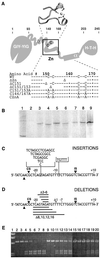

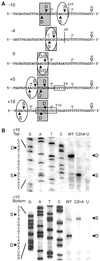
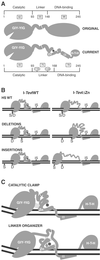
References
-
- Lambowitz A M, Belfort M. Annu Rev Biochem. 1993;62:587–622. - PubMed
-
- Lambowitz A M, Caprara M G, Zimmerly S, Perlman P S. In: The RNA World. Gesteland R F, Cech T R, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. pp. 451–485.
-
- Gimble F S. FEMS Microbiol Lett. 2000;185:99–107. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

