Association of a lysine-232/alanine polymorphism in a bovine gene encoding acyl-CoA:diacylglycerol acyltransferase (DGAT1) with variation at a quantitative trait locus for milk fat content
- PMID: 12077321
- PMCID: PMC123135
- DOI: 10.1073/pnas.142293799
Association of a lysine-232/alanine polymorphism in a bovine gene encoding acyl-CoA:diacylglycerol acyltransferase (DGAT1) with variation at a quantitative trait locus for milk fat content
Abstract
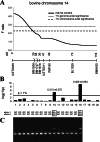
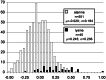
DGAT1 encodes diacylglycerol O-acyltransferase (EC ), a microsomal enzyme that catalyzes the final step of triglyceride synthesis. It became a functional candidate gene for lactation traits after studies indicated that mice lacking both copies of DGAT1 are completely devoid of milk secretion, most likely because of deficient triglyceride synthesis in the mammary gland. Our mapping studies placed DGAT1 close to the region of a quantitative trait locus (QTL) on bovine chromosome 14 for variation in fat content of milk. Sequencing of DGAT1 from pooled DNA revealed significant frequency shifts at several variable positions between groups of animals with high and low breeding values for milk fat content in different breeds (Holstein-Friesian, Fleckvieh, and Braunvieh). Among the variants was a nonconservative substitution of lysine by alanine (K232A), with the lysine-encoding allele being associated with higher milk fat content. Haplotype analysis indicated the lysine variant to be ancestral. Two animals that were typed heterozygous (Qq) at the QTL based on marker-assisted QTL-genotyping were heterozygous for the K232A substitution, whereas 14 animals that are most likely qq at the QTL were homozygous for the alanine-encoding allele. An independent association study in Fleckvieh animals confirmed the positive effect of the lysine variant on milk fat content. We consider the nonconservative K232A substitution to be directly responsible for the QTL variation, although our genetic studies cannot provide formal proof.
Figures




Similar articles
-
Effects of DGAT1 variants on milk production traits in German cattle breeds.J Anim Sci. 2003 Aug;81(8):1911-8. doi: 10.2527/2003.8181911x. J Anim Sci. 2003. PMID: 12926772
-
Evidence for multiple alleles at the DGAT1 locus better explains a quantitative trait locus with major effect on milk fat content in cattle.Genetics. 2004 Aug;167(4):1873-81. doi: 10.1534/genetics.103.022749. Genetics. 2004. PMID: 15342525 Free PMC article.
-
Characterization of the DGAT1 K232A and variable number of tandem repeat polymorphisms in French dairy cattle.J Dairy Sci. 2007 Jun;90(6):2980-8. doi: 10.3168/jds.2006-707. J Dairy Sci. 2007. PMID: 17517739
-
A mathematical model for mammary fatty acid synthesis and triglyceride assembly: the role of stearoyl CoA desaturase (SCD).J Dairy Res. 2004 Nov;71(4):385-97. doi: 10.1017/s0022029904000354. J Dairy Res. 2004. PMID: 15605704 Review.
-
Association of DGAT1 With Cattle, Buffalo, Goat, and Sheep Milk and Meat Production Traits.Front Vet Sci. 2021 Aug 16;8:712470. doi: 10.3389/fvets.2021.712470. eCollection 2021. Front Vet Sci. 2021. PMID: 34485439 Free PMC article. Review.
Cited by
-
The impact of genetic relationship information on genomic breeding values in German Holstein cattle.Genet Sel Evol. 2010 Feb 19;42(1):5. doi: 10.1186/1297-9686-42-5. Genet Sel Evol. 2010. PMID: 20170500 Free PMC article.
-
Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle.Nat Genet. 2014 Aug;46(8):858-65. doi: 10.1038/ng.3034. Epub 2014 Jul 13. Nat Genet. 2014. PMID: 25017103
-
Genome sequencing of the extinct Eurasian wild aurochs, Bos primigenius, illuminates the phylogeography and evolution of cattle.Genome Biol. 2015 Oct 26;16:234. doi: 10.1186/s13059-015-0790-2. Genome Biol. 2015. PMID: 26498365 Free PMC article.
-
Variant Amino Acid Residues Alter the Enzyme Activity of Peanut Type 2 Diacylglycerol Acyltransferases.Front Plant Sci. 2017 Oct 16;8:1751. doi: 10.3389/fpls.2017.01751. eCollection 2017. Front Plant Sci. 2017. PMID: 29085382 Free PMC article.
-
A physical map of the bovine genome.Genome Biol. 2007;8(8):R165. doi: 10.1186/gb-2007-8-8-r165. Genome Biol. 2007. PMID: 17697342 Free PMC article.
References
-
- Goddard M E, Wiggans G R. In: The Genetics of Cattle. Fries R, Ruvinsky A, editors. Wallingford, U.K.: CABI; 1999. pp. 511–537.
-
- Heyen D W, Weller J I, Ron M, Band M, Beever J E, Feldmesser E, Da Y, Wiggans G R, VanRanden P M, Lewin H A. Physiol Genomics. 1999;1:165–175. - PubMed
-
- Velmala R J, Vilkki H J, Elo K T, de Koning D J, Maki-Tanila A V. Anim Genet. 1999;30:136–143. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

