Drosophila Mi-2 negatively regulates dDREF by inhibiting its DNA-binding activity
- PMID: 12077345
- PMCID: PMC139784
- DOI: 10.1128/MCB.22.14.5182-5193.2002
Drosophila Mi-2 negatively regulates dDREF by inhibiting its DNA-binding activity
Abstract
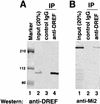
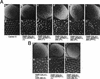
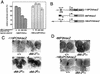
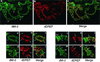
Drosophila melanogaster DNA replication-related element (DRE) factor (dDREF) is a transcriptional regulatory factor required for the expression of genes carrying the 5'-TATCGATA DRE. dDREF has been reported to bind to a sequence in the chromatin boundary element, and thus, dDREF may play a part in regulating insulator activity. To generate further insights into dDREF function, we carried out a Saccharomyces cerevisiae two-hybrid screening with DREF polypeptide as bait and identified Mi-2 as a DREF-interacting protein. Biochemical analyses revealed that the C-terminal region of Drosophila Mi-2 (dMi-2) specifically binds to the DNA-binding domain of dDREF. Electrophoretic mobility shift assays showed that dMi-2 thereby inhibits the DNA-binding activity of dDREF. Ectopic expression of dDREF and dMi-2 in eye imaginal discs resulted in severe and mild rough-eye phenotypes, respectively, whereas flies simultaneously expressing both proteins exhibited almost-normal eye phenotypes. Half-dose reduction of the dMi-2 gene enhanced the DREF-induced rough-eye phenotype. Immunostaining of polytene chromosomes of salivary glands showed that dDREF and dMi-2 bind in mutually exclusive ways. These lines of evidence define a novel function of dMi-2 in the negative regulation of dDREF by its DNA-binding activity. Finally, we postulated that dDREF and dMi-2 may demonstrate reciprocal regulation of their functions.
Figures



Similar articles
-
Drosophila Myc is required for normal DREF gene expression.Exp Cell Res. 2008 Jan 1;314(1):184-92. doi: 10.1016/j.yexcr.2007.09.014. Epub 2007 Sep 29. Exp Cell Res. 2008. PMID: 17963749
-
Drosophila distal-less negatively regulates dDREF by inhibiting its DNA binding activity.Biochim Biophys Acta. 2006 Jul;1759(7):359-66. doi: 10.1016/j.bbaexp.2006.07.002. Epub 2006 Jul 25. Biochim Biophys Acta. 2006. PMID: 16949685
-
Ectopic expression of DREF induces DNA synthesis, apoptosis, and unusual morphogenesis in the Drosophila eye imaginal disc: possible interaction with Polycomb and trithorax group proteins.Mol Cell Biol. 2001 Nov;21(21):7231-42. doi: 10.1128/MCB.21.21.7231-7242.2001. Mol Cell Biol. 2001. PMID: 11585906 Free PMC article.
-
The DRE/DREF transcriptional regulatory system: a master key for cell proliferation.Biochim Biophys Acta. 2008 Feb;1779(2):81-9. doi: 10.1016/j.bbagrm.2007.11.011. Epub 2007 Dec 4. Biochim Biophys Acta. 2008. PMID: 18155677 Review.
-
Damage control: the pleiotropy of DNA repair genes in Drosophila melanogaster.Genetics. 1998 Apr;148(4):1587-98. doi: 10.1093/genetics/148.4.1587. Genetics. 1998. PMID: 9560378 Free PMC article. Review. No abstract available.
Cited by
-
ZBED1/DREF: A transcription factor that regulates cell proliferation.Oncol Lett. 2020 Nov;20(5):137. doi: 10.3892/ol.2020.11997. Epub 2020 Aug 20. Oncol Lett. 2020. PMID: 32934705 Free PMC article. Review.
-
ATP-dependent chromatin remodeling complexes in Drosophila.Chromosome Res. 2006;14(4):433-49. doi: 10.1007/s10577-006-1067-0. Chromosome Res. 2006. PMID: 16821138 Review.
-
ATP-Dependent Chromatin Remodeling During Cortical Neurogenesis.Front Neurosci. 2018 Apr 9;12:226. doi: 10.3389/fnins.2018.00226. eCollection 2018. Front Neurosci. 2018. PMID: 29686607 Free PMC article. Review.
-
Arabidopsis chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate hypocotyl cell elongation.Plant Cell. 2013 Jan;25(1):242-56. doi: 10.1105/tpc.112.105742. Epub 2013 Jan 11. Plant Cell. 2013. PMID: 23314848 Free PMC article.
-
hDREF regulates cell proliferation and expression of ribosomal protein genes.Mol Cell Biol. 2007 Mar;27(6):2003-13. doi: 10.1128/MCB.01462-06. Epub 2007 Jan 12. Mol Cell Biol. 2007. PMID: 17220279 Free PMC article.
References
-
- Aubry, F., M.-G. Mattei, and F. Galibert. 1998. Identification of a human 17p-located cDNA encoding a protein of the Snf2-like helicase family. Eur. J. Biochem. 254:558-564. - PubMed
-
- Brand, A. H., and N. Perrimon. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401-415. - PubMed
-
- Duronio, R. J., P. H. O'Farrell, J. E. Xie, A. Brook, and N. Dyson. 1995. The transcription factor E2F is required for S phase during Drosophila embryogenesis. Genes Dev. 9:1445-1455. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
