Oxygen and nitrate-dependent regulation of dmsABC operon expression in Escherichia coli: sites for Fnr and NarL protein interactions
- PMID: 12079504
- PMCID: PMC116602
- DOI: 10.1186/1471-2180-2-13
Oxygen and nitrate-dependent regulation of dmsABC operon expression in Escherichia coli: sites for Fnr and NarL protein interactions
Abstract
Background: Escherichia coli, can respire anaerobically using dimethyl sulfoxide (DMSO) or trimethylamine-N-oxide (TMAO) as the terminal electron acceptor for anaerobic energy generation. Expression of the dmsABC genes that encode the membrane-associated DMSO/TMAO reductase is positively regulated during anaerobic conditions by the Fnr protein and negatively regulated by the NarL protein when nitrate is present.
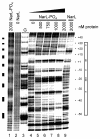
Results: The regions of dmsA regulatory DNA required for Fnr and NarL interactions in response to anaerobiosis and nitrate, respectively, were examined. Mutations within the Fnr site that deviated from the wild type sequence, TTGATaccgAACAA, or that removed an entire half-site, either impaired or abolished the anaerobic activation of dmsA-lacZ expression. The region for phosphorylated NarL (NarL-phosphate) binding at the dmsA promoter was identified by DNase I and hydroxyl radical footprinting methods. A large 97 bp region that overlaps the Fnr and RNA polymerase recognition sites was protected by NarL-phosphate but not by the non-phosphorylated form of NarL. Hydroxyl radical footprinting analysis confirmed the NarL-phosphate DNase I protections of both dmsA strands and revealed 8-9 protected sites of 3-5 bp occurring at ten bp intervals that are offset by 3 bp in the 3' direction.
Conclusion: These findings suggest that multiple molecules of phosphorylated NarL bind along one face of the DNA and may interfere with Fnr and/or RNA polymerase interactions at the dmsA regulatory region. The interplay of these transcription factors insures a hierarchical expression of the dmsABC genes when respiration of the preferred electron acceptors, oxygen and nitrate, is not possible.
Figures




Similar articles
-
Oxygen, nitrate, and molybdenum regulation of dmsABC gene expression in Escherichia coli.J Bacteriol. 1989 Jul;171(7):3817-23. doi: 10.1128/jb.171.7.3817-3823.1989. J Bacteriol. 1989. PMID: 2544558 Free PMC article.
-
Nitrate and nitrite regulation of the Fnr-dependent aeg-46.5 promoter of Escherichia coli K-12 is mediated by competition between homologous response regulators (NarL and NarP) for a common DNA-binding site.J Mol Biol. 1995 Aug 4;251(1):15-29. doi: 10.1006/jmbi.1995.0412. J Mol Biol. 1995. PMID: 7643383
-
Anaerobic regulation of the Escherichia coli dmsABC operon requires the molybdate-responsive regulator ModE.Mol Microbiol. 1998 Jan;27(1):197-208. doi: 10.1046/j.1365-2958.1998.00675.x. Mol Microbiol. 1998. PMID: 9466267
-
Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors.Biochim Biophys Acta. 1997 Jul 4;1320(3):217-34. doi: 10.1016/s0005-2728(97)00034-0. Biochim Biophys Acta. 1997. PMID: 9230919 Review.
-
Nitrate regulation of anaerobic respiratory gene expression in Escherichia coli.Mol Microbiol. 1993 Aug;9(3):425-34. doi: 10.1111/j.1365-2958.1993.tb01704.x. Mol Microbiol. 1993. PMID: 8412692 Review.
Cited by
-
Proteomic Analysis of FNR-Regulated Anaerobiosis in Salmonella Typhimurium.J Am Soc Mass Spectrom. 2019 Jun;30(6):1001-1012. doi: 10.1007/s13361-019-02145-2. Epub 2019 Mar 22. J Am Soc Mass Spectrom. 2019. PMID: 30903387
-
The coordinated action of RNase III and RNase G controls enolase expression in response to oxygen availability in Escherichia coli.Sci Rep. 2019 Nov 21;9(1):17257. doi: 10.1038/s41598-019-53883-y. Sci Rep. 2019. PMID: 31754158 Free PMC article.
-
Deinococcus geothermalis: the pool of extreme radiation resistance genes shrinks.PLoS One. 2007 Sep 26;2(9):e955. doi: 10.1371/journal.pone.0000955. PLoS One. 2007. PMID: 17895995 Free PMC article.
-
Iron limitation indirectly reduces the Escherichia coli torCAD operon expression by a reduction of molybdenum cofactor availability.Microbiol Spectr. 2024 Feb 6;12(2):e0348023. doi: 10.1128/spectrum.03480-23. Epub 2024 Jan 9. Microbiol Spectr. 2024. PMID: 38193660 Free PMC article.
-
UvrY is required for the full virulence of Aeromonas dhakensis.Virulence. 2020 Dec;11(1):502-520. doi: 10.1080/21505594.2020.1768339. Virulence. 2020. PMID: 32434424 Free PMC article.
References
-
- Bilous P, Cole S, Anderson W, Weiner J. Nucleotide sequence of the dmsABC operon encoding the anaerobic dimethylsulfoxide reductase of Escherichia coli. Mol Microbiol. 1988;2:785–795. - PubMed
-
- Albrecht J. Oxygen control of respiratory gene expression in Escherichia coli University of California at Los Angeles; 1996 PhD.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

