Characterization of HCP-6, a C. elegans protein required to prevent chromosome twisting and merotelic attachment
- PMID: 12080088
- PMCID: PMC186334
- DOI: 10.1101/gad.989102
Characterization of HCP-6, a C. elegans protein required to prevent chromosome twisting and merotelic attachment
Abstract
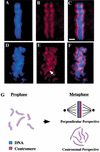
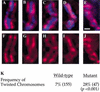
Previous studies of mitosis show that capture of single kinetochores by microtubules from both centrosomes (merotelic orientation) is a major cause of aneuploidy. We have characterized hcp-6, a temperature-sensitive chromosome segregation mutant in C. elegans that exhibits chromosomes attached to both poles via a single sister kinetochore. We demonstrate that the primary defect in this mutant is a failure to fully condense chromosomes during prophase. Although centromere formation and sister centromere resolution remain unaffected in hcp-6, the chromosomes lack the rigidity of wild-type chromosomes and twist around the long axis of the chromosome. As such, they are unable to establish a proper orientation at prometaphase, allowing individual kinetochores to be captured by microtubules from both poles. We therefore propose that chromosome rigidity plays an essential role in maintaining chromosome orientation to prevent merotelic capture.
Figures

Similar articles
-
Aurora B/AIR-2 regulates sister centromere resolution and CENP-A/HCP-3 organization to prevent merotelic attachments.J Mol Cell Biol. 2025 May 2;16(10):mjae045. doi: 10.1093/jmcb/mjae045. J Mol Cell Biol. 2025. PMID: 39415429 Free PMC article.
-
HCP-4/CENP-C promotes the prophase timing of centromere resolution by enabling the centromere association of HCP-6 in Caenorhabditis elegans.Mol Cell Biol. 2005 Apr;25(7):2583-92. doi: 10.1128/MCB.25.7.2583-2592.2005. Mol Cell Biol. 2005. PMID: 15767665 Free PMC article.
-
HCP-1, a protein involved in chromosome segregation, is localized to the centromere of mitotic chromosomes in Caenorhabditis elegans.J Cell Biol. 1999 Nov 1;147(3):471-80. doi: 10.1083/jcb.147.3.471. J Cell Biol. 1999. PMID: 10545493 Free PMC article.
-
Merotelic kinetochores in mammalian tissue cells.Philos Trans R Soc Lond B Biol Sci. 2005 Mar 29;360(1455):553-68. doi: 10.1098/rstb.2004.1610. Philos Trans R Soc Lond B Biol Sci. 2005. PMID: 15897180 Free PMC article. Review.
-
Merotelic kinetochore attachment: causes and effects.Trends Cell Biol. 2011 Jun;21(6):374-81. doi: 10.1016/j.tcb.2011.01.003. Epub 2011 Feb 8. Trends Cell Biol. 2011. PMID: 21306900 Free PMC article. Review.
Cited by
-
Condensins: universal organizers of chromosomes with diverse functions.Genes Dev. 2012 Aug 1;26(15):1659-78. doi: 10.1101/gad.194746.112. Genes Dev. 2012. PMID: 22855829 Free PMC article. Review.
-
Twenty years of merotelic kinetochore attachments: a historical perspective.Chromosome Res. 2023 Jul 19;31(3):18. doi: 10.1007/s10577-023-09727-7. Chromosome Res. 2023. PMID: 37466740 Free PMC article. Review.
-
An SMC-like protein binds and regulates Caenorhabditis elegans condensins.PLoS Genet. 2017 Mar 16;13(3):e1006614. doi: 10.1371/journal.pgen.1006614. eCollection 2017 Mar. PLoS Genet. 2017. PMID: 28301465 Free PMC article.
-
Timing of centrosome separation is important for accurate chromosome segregation.Mol Biol Cell. 2012 Feb;23(3):401-11. doi: 10.1091/mbc.E11-02-0095. Epub 2011 Nov 30. Mol Biol Cell. 2012. PMID: 22130796 Free PMC article.
-
Mitotic chromosome condensation requires phosphorylation of the centromeric protein KNL-2 in C. elegans.J Cell Sci. 2021 Dec 1;134(23):jcs259088. doi: 10.1242/jcs.259088. Epub 2021 Dec 2. J Cell Sci. 2021. PMID: 34734636 Free PMC article.
References
-
- Albertson DG, Thomson JN. The kinetochores of Caenorhabditis elegans. Chromosoma. 1982;86:409–428. - PubMed
-
- Buchwitz BJ, Ahmad K, Moore LL, Roth MB, Henikoff S. A histone-H3-like protein in C. elegans. Nature. 1999;401:547–548. - PubMed
-
- Cimini D, Fioravanti D, Salmon ED, Degrassi F. Merotelic kinetochore orientation versus chromosome mono-orientation in the origin of lagging chromosomes in human primary cells. J Cell Sci. 2002;115:507–515. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
