Targeted mutagenesis by homologous recombination in D. melanogaster
- PMID: 12080094
- PMCID: PMC186348
- DOI: 10.1101/gad.986602
Targeted mutagenesis by homologous recombination in D. melanogaster
Abstract
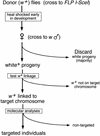
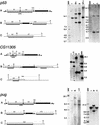
We used a recently developed method to produce mutant alleles of five endogenous Drosophila genes, including the homolog of the p53 tumor suppressor. Transgenic expression of the FLP site-specific recombinase and the I-SceI endonuclease generates extrachromosomal linear DNA molecules in vivo. These molecules undergo homologous recombination with the corresponding chromosomal locus to generate targeted alterations of the host genome. The results address several questions about the general utility of this technique. We show that genes not near telomeres can be efficiently targeted; that no knowledge of the mutant phenotype is needed for targeting; and that insertional mutations and allelic substitutions can be easily produced.
Figures





Similar articles
-
Gene targeting by homologous recombination in Drosophila.Science. 2000 Jun 16;288(5473):2013-8. doi: 10.1126/science.288.5473.2013. Science. 2000. PMID: 10856208
-
Ends-out, or replacement, gene targeting in Drosophila.Proc Natl Acad Sci U S A. 2003 Mar 4;100(5):2556-61. doi: 10.1073/pnas.0535280100. Epub 2003 Feb 14. Proc Natl Acad Sci U S A. 2003. PMID: 12589026 Free PMC article.
-
The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome.Cell. 1989 Nov 3;59(3):499-509. doi: 10.1016/0092-8674(89)90033-0. Cell. 1989. PMID: 2509077
-
Recombinases and their use in gene activation, gene inactivation, and transgenesis.Methods Mol Biol. 2008;420:175-95. doi: 10.1007/978-1-59745-583-1_10. Methods Mol Biol. 2008. PMID: 18641947 Review.
-
Gene-targeting and the p53 tumor-suppressor gene.Mutat Res. 1994 Jun 1;307(2):557-72. doi: 10.1016/0027-5107(94)90266-6. Mutat Res. 1994. PMID: 7514729 Review.
Cited by
-
Apoptotic mechanisms during competition of ribosomal protein mutant cells: roles of the initiator caspases Dronc and Dream/Strica.Cell Death Differ. 2015 Aug;22(8):1300-12. doi: 10.1038/cdd.2014.218. Epub 2015 Jan 23. Cell Death Differ. 2015. PMID: 25613379 Free PMC article.
-
Efficient gene targeting in Drosophila with zinc-finger nucleases.Genetics. 2006 Apr;172(4):2391-403. doi: 10.1534/genetics.105.052829. Epub 2006 Feb 1. Genetics. 2006. PMID: 16452139 Free PMC article.
-
Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage.Mol Cell Biol. 2004 Feb;24(3):1219-31. doi: 10.1128/MCB.24.3.1219-1231.2004. Mol Cell Biol. 2004. PMID: 14729967 Free PMC article.
-
Drosophila p53 preserves genomic stability by regulating cell death.Proc Natl Acad Sci U S A. 2003 Apr 15;100(8):4696-701. doi: 10.1073/pnas.0736384100. Epub 2003 Apr 2. Proc Natl Acad Sci U S A. 2003. PMID: 12672954 Free PMC article.
-
Stereotypic founder cell patterning and embryonic muscle formation in Drosophila require nautilus (MyoD) gene function.Proc Natl Acad Sci U S A. 2007 Mar 27;104(13):5461-6. doi: 10.1073/pnas.0608739104. Epub 2007 Mar 21. Proc Natl Acad Sci U S A. 2007. PMID: 17376873 Free PMC article.
References
-
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. - PubMed
-
- Anonymous. In search of a function. Nat Cell Biol. 2000;2:E137–E138. - PubMed
-
- Argast GM, Stephens KM, Emond MJ, Monnat RJ. I-PpoI and I-CreI homing site sequence degeneracy determined by random mutagenesis and sequential in vitro enrichment. J Mol Biol. 1998;280:345–353. - PubMed
-
- Bollag RJ, Waldman AS, Liskay RM. Homologous recombination in mammalian cells. Ann Rev Genet. 1989;23:199–225. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
