Positional cloning of the murine flavivirus resistance gene
- PMID: 12080145
- PMCID: PMC123139
- DOI: 10.1073/pnas.142287799
Positional cloning of the murine flavivirus resistance gene
Abstract
Inbred mouse strains exhibit significant differences in their susceptibility to viruses in the genus Flavivirus, which includes human pathogens such as yellow fever, Dengue, and West Nile virus. A single gene, designated Flv, confers this differential susceptibility and was mapped previously to a region of mouse chromosome 5. A positional cloning strategy was used to identify 22 genes from the Flv gene interval including 10 members of the 2'-5'-oligoadenylate synthetase gene family. One 2'-5'-oligoadenylate synthetase gene, Oas1b, was identified as Flv by correlation between genotype and phenotype in nine mouse strains. Susceptible mouse strains produce a protein lacking 30% of the C-terminal sequence as compared with the resistant counterpart because of the presence of a premature stop codon. The Oas1b gene differs from all the other murine Oas genes by a unique four-amino acid deletion in the P-loop located within the conserved RNA binding domain. Expression of the resistant allele of Oas1b in susceptible embryo fibroblasts resulted in partial inhibition of the replication of a flavivirus but not of an alpha togavirus.
Figures


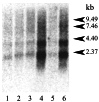
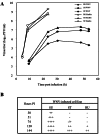

Comment in
-
Host genetic variability and West Nile virus susceptibility.Proc Natl Acad Sci U S A. 2002 Sep 3;99(18):11555-7. doi: 10.1073/pnas.202448899. Epub 2002 Aug 21. Proc Natl Acad Sci U S A. 2002. PMID: 12192094 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

