Export of L-isoleucine from Corynebacterium glutamicum: a two-gene-encoded member of a new translocator family
- PMID: 12081967
- PMCID: PMC135157
- DOI: 10.1128/JB.184.14.3947-3956.2002
Export of L-isoleucine from Corynebacterium glutamicum: a two-gene-encoded member of a new translocator family
Abstract
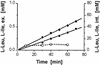
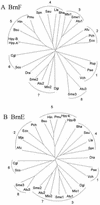
Bacteria possess amino acid export systems, and Corynebacterium glutamicum excretes L-isoleucine in a process dependent on the proton motive force. In order to identify the system responsible for L-isoleucine export, we have used transposon mutagenesis to isolate mutants of C. glutamicum sensitive to the peptide isoleucyl-isoleucine. In one such mutant, strong peptide sensitivity resulted from insertion into a gene designated brnF encoding a hydrophobic protein predicted to possess seven transmembrane spanning helices. brnE is located downstream of brnF and encodes a second hydrophobic protein with four putative membrane-spanning helices. A mutant deleted of both genes no longer exports L-isoleucine, whereas an overexpressing strain exports this amino acid at an increased rate. BrnF and BrnE together are also required for the export of L-leucine and L-valine. BrnFE is thus a two-component export permease specific for aliphatic hydrophobic amino acids. Upstream of brnFE and transcribed divergently is an Lrp-like regulatory gene required for active export. Searches for homologues of BrnFE show that this type of exporter is widespread in prokaryotes but lacking in eukaryotes and that both gene products which together comprise the members of a novel family, the LIV-E family, generally map together within a single operon. Comparisons of the BrnF and BrnE phylogenetic trees show that gene duplication events in the early bacterial lineage gave rise to multiple paralogues that have been retained in alpha-proteobacteria but not in other prokaryotes analyzed.
Figures


Similar articles
-
Lrp of Corynebacterium glutamicum controls expression of the brnFE operon encoding the export system for L-methionine and branched-chain amino acids.J Biotechnol. 2012 Apr 30;158(4):231-41. doi: 10.1016/j.jbiotec.2011.06.003. Epub 2011 Jun 12. J Biotechnol. 2012. PMID: 21683740
-
Characterization of methionine export in Corynebacterium glutamicum.J Bacteriol. 2005 Jun;187(11):3786-94. doi: 10.1128/JB.187.11.3786-3794.2005. J Bacteriol. 2005. PMID: 15901702 Free PMC article.
-
Increasing l-isoleucine production in Corynebacterium glutamicum by overexpressing global regulator Lrp and two-component export system BrnFE.J Appl Microbiol. 2013 May;114(5):1369-77. doi: 10.1111/jam.12141. Epub 2013 Feb 8. J Appl Microbiol. 2013. PMID: 23331988
-
New ubiquitous translocators: amino acid export by Corynebacterium glutamicum and Escherichia coli.Arch Microbiol. 2003 Sep;180(3):155-60. doi: 10.1007/s00203-003-0581-0. Epub 2003 Jul 15. Arch Microbiol. 2003. PMID: 12879215 Review.
-
Molecular aspects of lysine, threonine, and isoleucine biosynthesis in Corynebacterium glutamicum.Antonie Van Leeuwenhoek. 1993-1994;64(2):145-63. doi: 10.1007/BF00873024. Antonie Van Leeuwenhoek. 1993. PMID: 8092856 Review.
Cited by
-
MtcB, a member of the MttB superfamily from the human gut acetogen Eubacterium limosum, is a cobalamin-dependent carnitine demethylase.J Biol Chem. 2020 Aug 21;295(34):11971-11981. doi: 10.1074/jbc.RA120.012934. Epub 2020 Jun 22. J Biol Chem. 2020. PMID: 32571881 Free PMC article.
-
Microbial methionine transporters and biotechnological applications.Appl Microbiol Biotechnol. 2021 May;105(10):3919-3929. doi: 10.1007/s00253-021-11307-w. Epub 2021 Apr 30. Appl Microbiol Biotechnol. 2021. PMID: 33929594 Free PMC article. Review.
-
CRISPR-assisted rational flux-tuning and arrayed CRISPRi screening of an L-proline exporter for L-proline hyperproduction.Nat Commun. 2022 Feb 16;13(1):891. doi: 10.1038/s41467-022-28501-7. Nat Commun. 2022. PMID: 35173152 Free PMC article.
-
Characterization and molecular mechanism of AroP as an aromatic amino acid and histidine transporter in Corynebacterium glutamicum.J Bacteriol. 2013 Dec;195(23):5334-42. doi: 10.1128/JB.00971-13. Epub 2013 Sep 20. J Bacteriol. 2013. PMID: 24056108 Free PMC article.
-
Application of a genetically encoded biosensor for live cell imaging of L-valine production in pyruvate dehydrogenase complex-deficient Corynebacterium glutamicum strains.PLoS One. 2014 Jan 17;9(1):e85731. doi: 10.1371/journal.pone.0085731. eCollection 2014. PLoS One. 2014. PMID: 24465669 Free PMC article.
References
-
- Bellmann, A., M. Vrljic, M. Pátek, H. Sahm, R. Krämer, and L. Eggeling. 2001. Expression control and specificity of the basic amino acid exporter LysE of Corynebacterium glutamicum. Microbiology 147:1765-1774. - PubMed
-
- Chung, Y. J., and M. H. Saier, Jr. 2001. SMR-type multidrug resistance pumps. Curr. Opin. Drug Dis. Dev. 4:237-245. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

