The genome-wide expression response to telomerase deletion in Saccharomyces cerevisiae
- PMID: 12084816
- PMCID: PMC123138
- DOI: 10.1073/pnas.142162499
The genome-wide expression response to telomerase deletion in Saccharomyces cerevisiae
Erratum in
- Proc Natl Acad Sci U S A. 2004 Aug 31;101(35):13100
Abstract
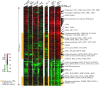
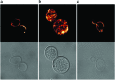
Loss of the protective function of telomeres has previously been hypothesized to cause a DNA damage response. Here, we report a genome-wide expression response, the telomerase deletion response (TDR), that occurs when telomeres can no longer be maintained by telomerase. The TDR shares features with other DNA damage responses and the environmental stress response. Unexpectedly, another feature of the TDR is the up-regulation of energy production genes, accompanied by a proliferation of mitochondria. Finally, a discrete set of genes, the "telomerase deletion signature", is uniquely up-regulated in the TDR but not under other conditions of stress and DNA damage that have been reported. The telomerase deletion signature genes define new candidates for involvement in cellular responses to altered telomere structure or function.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

