Contrapuntal networks of gene expression during Arabidopsis seed filling
- PMID: 12084821
- PMCID: PMC150774
- DOI: 10.1105/tpc.000877
Contrapuntal networks of gene expression during Arabidopsis seed filling
Abstract
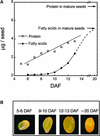
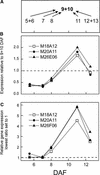
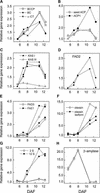
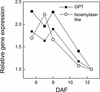
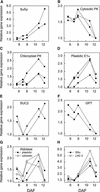
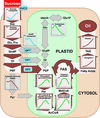
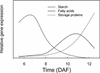
We have used cDNA microarrays to examine changes in gene expression during Arabidopsis seed development and to compare wild-type and mutant wrinkled1 (wri1) seeds that have an 80% reduction in oil. Between 5 and 13 days after flowering, a period preceding and including the major accumulation of storage oils and proteins, approximately 35% of the genes represented on the array changed at least twofold, but a larger fraction (65%) showed little or no change in expression. Genes whose expression changed most tended to be expressed more in seeds than in other tissues. Genes related to the biosynthesis of storage components showed several distinct temporal expression patterns. For example, a number of genes encoding core fatty acid synthesis enzymes displayed a bell-shaped pattern of expression between 5 and 13 days after flowering. By contrast, the expression of storage proteins, oleosins, and other known abscisic acid-regulated genes increased later and remained high. Genes for photosynthetic proteins followed a pattern very similar to that of fatty acid synthesis proteins, implicating a role in CO(2) refixation and the supply of cofactors for oil synthesis. Expression profiles of key carbon transporters and glycolytic enzymes reflected shifts in flux from cytosolic to plastid metabolism. Despite major changes in metabolism between wri1 and wild-type seeds, <1% of genes differed by more than twofold, and most of these were involved in central lipid and carbohydrate metabolism. Thus, these data define in part the downstream responses to disruption of the WRI1 gene.
Figures

References
-
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. - PubMed
-
- Bowman, J.L., and Mansfield, S.G. (1994). Embryogenesis: Introduction. In Arabidopsis: An Atlas of Morphology and Development, J.L. Bowman, ed (New York: Springer-Verlag), pp. 351–361.
-
- Browse, J., McCourt, P.J., and Somerville, C.R. (1986). Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Anal. Biochem. 152, 141–145. - PubMed
-
- DeRisi, J.L., Iyer, V.R., and Brown, P.O. (1997). Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278, 680–686. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

