In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis
- PMID: 12084833
- PMCID: PMC150786
- DOI: 10.1105/tpc.001628
In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis
Abstract
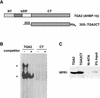
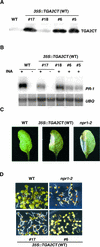
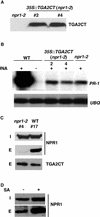
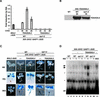
The Arabidopsis NPR1 protein is a key regulator of salicylic acid (SA)-mediated gene expression in systemic acquired resistance. Based on yeast two-hybrid analysis, NPR1 has been suggested to interact with members of the TGA family of transcription factors, including TGA2 (AHBP-1b). However, genetic evidence demonstrating that the NPR1-TGA interaction occurs in planta is still lacking, and the role of this interaction in SA-mediated gene activation has yet to be determined. In this study, we expressed a truncated form of TGA2 in Arabidopsis and found that the resulting transgenic lines displayed phenotypes similar to those of npr1 mutants. This dominant-negative effect of the TGA2 mutant shows that TGA2 and NPR1 interact in planta. We also present biochemical evidence indicating that this interaction is specific and enhanced by SA treatment. Moreover, using a chimera reporter system, we found that a chimeric TGA2GAL4 transcription factor activated a UAS(GAL)::GUS reporter gene in response to SA and that this activation was abolished in the npr1 mutant. NPR1 is required for the DNA binding activity of the transcription factor. These genetic data clearly demonstrate that TGA2 is a SA-responsive and NPR1-dependent transcription activator.
Figures


Similar articles
-
Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance.Plant Cell. 2003 Nov;15(11):2647-53. doi: 10.1105/tpc.014894. Epub 2003 Oct 23. Plant Cell. 2003. PMID: 14576289 Free PMC article.
-
Over-expression of TGA5, which encodes a bZIP transcription factor that interacts with NIM1/NPR1, confers SAR-independent resistance in Arabidopsis thaliana to Peronospora parasitica.Plant J. 2002 Oct;32(2):151-63. doi: 10.1046/j.1365-313x.2001.01411.x. Plant J. 2002. PMID: 12383081
-
NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid.Mol Plant Microbe Interact. 2000 Feb;13(2):191-202. doi: 10.1094/MPMI.2000.13.2.191. Mol Plant Microbe Interact. 2000. PMID: 10659709
-
NPR1, all things considered.Curr Opin Plant Biol. 2004 Oct;7(5):547-52. doi: 10.1016/j.pbi.2004.07.005. Curr Opin Plant Biol. 2004. PMID: 15337097 Review.
-
NPR1: the spider in the web of induced resistance signaling pathways.Curr Opin Plant Biol. 2004 Aug;7(4):456-64. doi: 10.1016/j.pbi.2004.05.006. Curr Opin Plant Biol. 2004. PMID: 15231270 Review.
Cited by
-
Microbial effectors target multiple steps in the salicylic acid production and signaling pathway.Front Plant Sci. 2015 May 19;6:349. doi: 10.3389/fpls.2015.00349. eCollection 2015. Front Plant Sci. 2015. PMID: 26042138 Free PMC article. Review.
-
The NPR1 homolog GhNPR1 plays an important role in the defense response of Gladiolus hybridus.Plant Cell Rep. 2015 Jun;34(6):1063-74. doi: 10.1007/s00299-015-1765-1. Epub 2015 Feb 24. Plant Cell Rep. 2015. PMID: 25708873
-
Diverse Roles of the Salicylic Acid Receptors NPR1 and NPR3/NPR4 in Plant Immunity.Plant Cell. 2020 Dec;32(12):4002-4016. doi: 10.1105/tpc.20.00499. Epub 2020 Oct 9. Plant Cell. 2020. PMID: 33037144 Free PMC article.
-
NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol.Plant Cell. 2003 Mar;15(3):760-70. doi: 10.1105/tpc.009159. Plant Cell. 2003. PMID: 12615947 Free PMC article.
-
Improved Powdery Mildew Resistance of Transgenic Nicotiana benthamiana Overexpressing the Cucurbita moschata CmSGT1 Gene.Front Plant Sci. 2019 Jul 25;10:955. doi: 10.3389/fpls.2019.00955. eCollection 2019. Front Plant Sci. 2019. PMID: 31402923 Free PMC article.
References
-
- Aoyama, T., and Chua, N.-H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11, 605–612. - PubMed
-
- Aravind, L., and Koonin, E.V. (1999). Fold prediction and evolutionary analysis of the POZ domain: Structural and evolutionary relationship with the potassium channel tetramerization domain. J. Mol. Biol. 285, 1353–1361. - PubMed
-
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

