The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis
- PMID: 12084834
- PMCID: PMC150787
- DOI: 10.1105/tpc.000869
The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis
Abstract
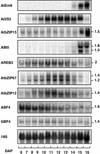
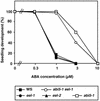
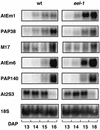
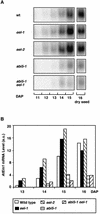
In Arabidopsis, the basic leucine zipper transcription factor ABI5 activates several late embryogenesis-abundant genes, including AtEm1 and AtEm6. However, the expression of many other seed maturation genes is independent of ABI5. We investigated the possibility that ABI5 homologs also participate in the regulation of gene expression during seed maturation. We identified 13 ABI5-related genes in the Arabidopsis genomic sequence. RNA gel blot analysis showed that seven of these genes are active during seed maturation and that they display distinct expression kinetics. We isolated and characterized two mutant alleles of one of these genes, AtbZIP12/EEL. Unlike abi5, the eel mutations did not inhibit the expression of any of the maturation marker genes that we monitored. On the contrary, the accumulation of the AtEm1 and AtEm6 mRNAs was enhanced in eel mutant seeds compared with wild-type seeds. Gel mobility shift assays, combined with analysis of the genetic interactions among the eel and abi5 mutations, indicated that ABI5 and EEL compete for the same binding sites within the AtEm1 promoter. This study illustrates how two homologous transcription factors can play antagonistic roles to fine-tune gene expression.
Figures


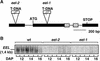

References
-
- Asamizu, E., Nakamura, Y., Sato, S., and Tabata, S. (2000). A large scale analysis of cDNA in Arabidopsis thaliana: Generation of 12,028 non-redundant expressed sequence tags from normalized and size-selected cDNA libraries. DNA Res. 7, 175–180. - PubMed
-
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. 316, 1194–1199. - PubMed
-
- Choi, H., Hong, J., Ha, J., Kang, J., and Kim, S.Y. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275, 1723–1730. - PubMed
-
- Delseny, M., Bies-Etheve, N., Carles, C., Hull, G., Vicient, C.M., Raynal, M., Grellet, F., and Aspart, L. (2001). Late embryogenesis abundant (LEA) protein gene regulation during Arabidopsis seed maturation. J. Plant Physiol. 158, 419–427.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

