Omnipotent decoding potential resides in eukaryotic translation termination factor eRF1 of variant-code organisms and is modulated by the interactions of amino acid sequences within domain 1
- PMID: 12084909
- PMCID: PMC124286
- DOI: 10.1073/pnas.142690099
Omnipotent decoding potential resides in eukaryotic translation termination factor eRF1 of variant-code organisms and is modulated by the interactions of amino acid sequences within domain 1
Abstract
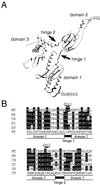
In eukaryotes, a single translational release factor, eRF1, deciphers three stop codons, although its decoding mechanism remains puzzling. In the ciliate Tetrahymena thermophila, UAA and UAG codons are reassigned to Gln codons. A yeast eRF1-domain swap containing Tetrahymena domain 1 responded only to UGA in vitro and failed to complement a defect in yeast eRF1 in vivo at 37 degrees C. This finding demonstrates that decoding specificity of eRF1 from variant code organisms resides at domain 1. However, the wild-type eRF1 hybrid fully restored the growth of eRF1-deficient yeast at 30 degrees C. Tetrahymena eRF1 contains a variant sequence, KATNIKD, at the tip of domain 1. The TASNIKD variant of hybrid eRF1 rendered the eRF1-nullified yeast viable, although in an in vitro assay, the same hybrid eRF1 responded only to UGA. Nevertheless, the yeast eRF1 bearing the KATNIKD motif instead of the TASNIKS heptapeptide present in higher eukaryotes remains omnipotent in vivo. Collectively, these data suggest that variant genetic code organisms like Tetrahymena have an intrinsic potential to decode three stop codons in vivo, and that interaction within domain 1 between the KAT tripeptide and other sequences modulates the decoding specificity of Tetrahymena eRF1.
Figures


References
-
- Nakamura Y, Ito K, Ehrenberg M. Cell. 2000;101:349–352. - PubMed
-
- Kisselev L L, Buckingham R H. Trends Biochem Sci. 2000;25:561–567. - PubMed
-
- Nakamura Y, Ito K, Isaksson L A. Cell. 1996;87:147–150. - PubMed
-
- Tate W P, Poole E S, Mannering S A. Prog Nucleic Acids Res. 1996;52:293–335. - PubMed
-
- Ito K, Uno M, Nakamura Y. Nature (London) 2000;403:680–684. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

